Efficient Production of On-Target Reads for Small RNA Sequencing of Single Cells Using Modified Adapters
- PMID: 30260208
- PMCID: PMC6233959
- DOI: 10.1021/acs.analchem.8b02773
Efficient Production of On-Target Reads for Small RNA Sequencing of Single Cells Using Modified Adapters
Abstract
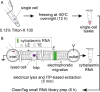
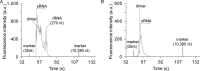
Although single-cell mRNA sequencing has been a powerful tool to explore cellular heterogeneity, the sequencing of small RNA at the single-cell level (sc-sRNA-seq) remains a challenge, as these have no consensus sequence, are relatively low abundant, and are difficult to amplify in a bias-free fashion. We present two methods of single-cell-lysis that enable sc-sRNA-seq. The first method is a chemical-based technique with overnight freezing while the second method leverages on-chip electrical lysis of plasma membrane and physical extraction and separation of cytoplasmic RNA via isotachophoresis. We coupled these two methods with off-chip small RNA library preparation using CleanTag modified adapters to prevent the formation of adapter dimers. We then demonstrated sc-sRNA-seq with single K562 human leukemic cells. Our approaches offer a relatively short hands-on time of 6 h and efficient generation of on-target reads. The sc-sRNA-seq with our approaches showed detection of miRNA with various abundances ranging from 16 000 copies/cell to about 10 copies/cell. We anticipate this approach will create a new opportunity to explore cellular heterogeneity through small RNA expression.
Conflict of interest statement
The authors declare no competing financial interest.
Figures
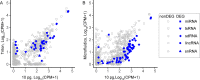
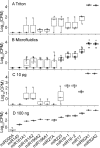
Similar articles
-
CleanTag Adapters Improve Small RNA Next-Generation Sequencing Library Preparation by Reducing Adapter Dimers.Methods Mol Biol. 2018;1712:145-161. doi: 10.1007/978-1-4939-7514-3_10. Methods Mol Biol. 2018. PMID: 29224073
-
Small RNA Profiling by Next-Generation Sequencing Using High-Definition Adapters.Methods Mol Biol. 2017;1580:45-57. doi: 10.1007/978-1-4939-6866-4_4. Methods Mol Biol. 2017. PMID: 28439825
-
Small RNA Profiling by Next-Generation Sequencing Using High-Definition Adapters.Methods Mol Biol. 2023;2630:103-115. doi: 10.1007/978-1-0716-2982-6_8. Methods Mol Biol. 2023. PMID: 36689179
-
Biases in small RNA deep sequencing data.Nucleic Acids Res. 2014 Feb;42(3):1414-26. doi: 10.1093/nar/gkt1021. Epub 2013 Nov 5. Nucleic Acids Res. 2014. PMID: 24198247 Free PMC article. Review.
-
[Advance in Deep Sequencing of Small RNAs for Virus Identification and Discovery].Bing Du Xue Bao. 2015 Jul;31(4):457-62. Bing Du Xue Bao. 2015. PMID: 26524920 Review. Chinese.
Cited by
-
Liver in infections: a single-cell and spatial transcriptomics perspective.J Biomed Sci. 2023 Jul 10;30(1):53. doi: 10.1186/s12929-023-00945-z. J Biomed Sci. 2023. PMID: 37430371 Free PMC article. Review.
-
NanoSINC-seq dissects the isoform diversity in subcellular compartments of single cells.Sci Adv. 2021 Apr 7;7(15):eabe0317. doi: 10.1126/sciadv.abe0317. Print 2021 Apr. Sci Adv. 2021. PMID: 33827812 Free PMC article.
-
Simultaneous RNA purification and size selection using on-chip isotachophoresis with an ionic spacer.Lab Chip. 2019 Aug 21;19(16):2741-2749. doi: 10.1039/c9lc00311h. Epub 2019 Jul 22. Lab Chip. 2019. PMID: 31328753 Free PMC article.
-
Inferring single-cell and spatial microRNA activity from transcriptomics data.Commun Biol. 2025 Jan 18;8(1):87. doi: 10.1038/s42003-025-07454-9. Commun Biol. 2025. PMID: 39827321 Free PMC article.
-
A SINC-Seq Protocol for the Analysis of Subcellular Gene Expression in Single Cells.Methods Mol Biol. 2023;2689:179-189. doi: 10.1007/978-1-0716-3323-6_14. Methods Mol Biol. 2023. PMID: 37430055
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

