RNF138 confers cisplatin resistance in gastric cancer cells via activating Chk1 signaling pathway
- PMID: 30260263
- PMCID: PMC6301830
- DOI: 10.1080/15384047.2018.1480293
RNF138 confers cisplatin resistance in gastric cancer cells via activating Chk1 signaling pathway
Erratum in
-
Correction.Cancer Biol Ther. 2024 Dec 31;25(1):2375109. doi: 10.1080/15384047.2024.2375109. Epub 2024 Jul 1. Cancer Biol Ther. 2024. PMID: 38951516 Free PMC article. No abstract available.
Abstract
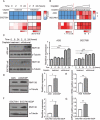
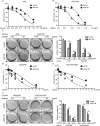
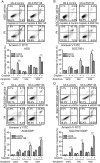
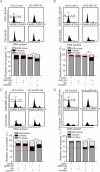
Chemotherapy resistance represents a major issue associated with gastric cancer (GC) treatment, and arises through multiple mechanisms, including modulation of the cell-cycle check point. Several ubiquitin kinases, including RING finger protein 138 (RNF138), have been reported to mediate the G2/M phase arrest. In this study, we investigated the role of RNF138 in the development of cisplatin resistance of two GC cell lines. We show that RNF138 levels are higher in cisplatin-resistant cell lines, compared with cisplatin-sensitive cells, and RNF138 expression was elevated during drug withdrawal following the cisplatin treatment. Using gene overexpression and silencing, we analyzed the impact of altering RNF138 level on GC cell viability, apoptosis, and cell cycle phenotypes in two isogenic cisplatin-sensitive and resistant cell lines. We show that RNF138 overexpression increased GC cell viability, decreased apoptosis and delayed cell cycle progression in the cisplatin-sensitive GC cells. Conversely, RNF138 silencing produced opposite phenotypes in the cisplatin-resistant cells. Moreover, RNF138-dependent phosphorylation of Chk1 was seen in GC cells, indicating a novel connection between cisplatin-induced DNA damage and apoptosis. Collectively, these data suggest that RNF138 modulates the cisplatin resistance in the GC cells, thus serving as a potential drug target to challenge chemotherapy failure. In addition, RNF138 can also be used as a marker to monitor the development of cisplatin resistance in GC treatment.
Keywords: Chk1; RNF138; apoptosis; cell cycle; cisplatin resistance; gastric cancer; viability.
Figures

Similar articles
-
RNF138 contributes to cisplatin resistance in nasopharyngeal carcinoma cells.Sci Rep. 2025 Jan 9;15(1):1406. doi: 10.1038/s41598-025-85716-6. Sci Rep. 2025. PMID: 39789198 Free PMC article.
-
Increasing cisplatin sensitivity by schedule-dependent inhibition of AKT and Chk1.Cancer Biol Ther. 2014;15(12):1600-12. doi: 10.4161/15384047.2014.961876. Cancer Biol Ther. 2014. PMID: 25482935 Free PMC article.
-
Downregulation of Inhibition of Apoptosis-Stimulating Protein of p53 (iASPP) Suppresses Cisplatin-Resistant Gastric Carcinoma In Vitro.Med Sci Monit. 2017 Nov 21;23:5542-5549. doi: 10.12659/msm.905403. Med Sci Monit. 2017. PMID: 29161238 Free PMC article.
-
Inhibition of AKT sensitizes chemoresistant ovarian cancer cells to cisplatin by abrogating S and G2/M arrest.Exp Mol Pathol. 2016 Jun;100(3):506-13. doi: 10.1016/j.yexmp.2016.05.003. Epub 2016 May 6. Exp Mol Pathol. 2016. PMID: 27163202
-
Cisplatin resistance: a cellular self-defense mechanism resulting from multiple epigenetic and genetic changes.Pharmacol Rev. 2012 Jul;64(3):706-21. doi: 10.1124/pr.111.005637. Epub 2012 Jun 1. Pharmacol Rev. 2012. PMID: 22659329 Free PMC article. Review.
Cited by
-
Mechanism of cisplatin resistance in gastric cancer and associated microRNAs.Cancer Chemother Pharmacol. 2023 Nov;92(5):329-340. doi: 10.1007/s00280-023-04572-1. Epub 2023 Aug 3. Cancer Chemother Pharmacol. 2023. PMID: 37535106 Review.
-
Silencing microRNA‑29b‑3p expression protects human trabecular meshwork cells against oxidative injury via upregulation of RNF138 to activate the ERK pathway.Int J Mol Med. 2021 Jun;47(6):101. doi: 10.3892/ijmm.2021.4934. Epub 2021 Apr 28. Int J Mol Med. 2021. PMID: 33907817 Free PMC article.
-
RNF138 Downregulates Antiviral Innate Immunity by Inhibiting IRF3 Activation.Int J Mol Sci. 2023 Nov 9;24(22):16110. doi: 10.3390/ijms242216110. Int J Mol Sci. 2023. PMID: 38003298 Free PMC article.
-
E3 Ubiquitin Ligase in Anticancer Drugdsla Resistance: Recent Advances and Future Potential.Front Pharmacol. 2021 Apr 15;12:645864. doi: 10.3389/fphar.2021.645864. eCollection 2021. Front Pharmacol. 2021. PMID: 33935743 Free PMC article. Review.
-
LncRNA HLA Complex Group 11 Knockdown Alleviates Cisplatin Resistance in Gastric Cancer by Targeting the miR-144-3p/UBE2D1 Axis.Cancer Manag Res. 2021 Oct 1;13:7543-7557. doi: 10.2147/CMAR.S329846. eCollection 2021. Cancer Manag Res. 2021. Retraction in: Cancer Manag Res. 2023 Sep 05;15:979-980. doi: 10.2147/CMAR.S438016. PMID: 34629901 Free PMC article. Retracted.
References
-
- Zhao L, Pan Y, Gang Y, Wang H, Jin H, Tie J, Xia L, Zhang Y, He L, Yao L, et al. Identification of GAS1 as an epirubicin resistance-related gene in human gastric cancer cells with a partially randomized small interfering RNA library. J Biol Chem. 2009; 284(39): 26273–26285.doi:10.1074/jbc.M109.028068. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
