Differential escape of HCV from CD8+ T cell selection pressure between China and Germany depends on the presenting HLA class I molecule
- PMID: 30260541
- PMCID: PMC7379502
- DOI: 10.1111/jvh.13011
Differential escape of HCV from CD8+ T cell selection pressure between China and Germany depends on the presenting HLA class I molecule
Abstract
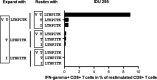
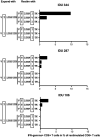
Adaptation of hepatitis C virus (HCV) to CD8+ T cell selection pressure is well described; however, it is unclear if HCV differentially adapts in different populations. Here, we studied HLA class I-associated viral sequence polymorphisms in HCV 1b isolates in a Chinese population and compared viral substitution patterns between Chinese and German populations. We identified three HLA class I-restricted epitopes in HCV NS3 with statistical support for selection pressure and found evidence for differential escape pathways between isolates from China and Germany depending on the HLA class I molecule. The substitution patterns particularly differed in the epitope VTLTHPITK1635-1643 , which was presented by HLA-A*03 as well as HLA-A*11, two alleles with highly different frequencies in the two populations. In Germany, a substitution in position seven of the epitope was the most frequent substitution in the presence of HLA-A*03, functionally associated with immune escape and nearly absent in Chinese isolates. In contrast, the most frequent substitution in China was located at position two of the epitope and became the predominant consensus residue. Moreover, substitutions in position one of the epitope were significantly enriched in HLA-A*11-positive individuals in China and associated with different patterns of CD8+ T cell reactivity. Our study confirms the differential escape pathways selected by HCV that depended on different HLA class I alleles in Chinese and German populations, indicating that HCV differentially adapts to distinct HLA class I alleles in these populations. This result has important implications for vaccine design against highly variable and globally distributed pathogens, which may require matching antigen sequences to geographic regions for T cell-based vaccine strategies.
© 2018 The Authors. Journal of Viral Hepatitis Published by John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare there is no conflict of interest regarding the publication of this paper.
Figures

References
-
- WHO . Hepatitis C. 2017. http://www.who.int/mediacentre/factsheets/fs164/en/. Accessed January 25, 2018.
-
- Asselah T, Boyer N, Saadoun D, Martinotpeignoux M, Marcellin P. Direct‐acting antivirals for the treatment of hepatitis C virus infection: optimizing current IFN‐free treatment and future perspectives. Liver Int. 2016;36(Suppl. 1):47‐57. - PubMed
-
- Grüner NH, Gerlach TJ, Jung M‐C, et al. Association of hepatitis C virus—specific CD8 + T cells with viral clearance in acute hepatitis C. J Infect Dis. 2000;181:1528‐1536. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

