Genetic diversity, infection prevalence, and possible transmission routes of Bartonella spp. in vampire bats
- PMID: 30260954
- PMCID: PMC6159870
- DOI: 10.1371/journal.pntd.0006786
Genetic diversity, infection prevalence, and possible transmission routes of Bartonella spp. in vampire bats
Abstract
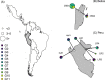
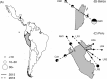
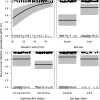
Bartonella spp. are globally distributed bacteria that cause endocarditis in humans and domestic animals. Recent work has suggested bats as zoonotic reservoirs of some human Bartonella infections; however, the ecological and spatiotemporal patterns of infection in bats remain largely unknown. Here we studied the genetic diversity, prevalence of infection across seasons and years, individual risk factors, and possible transmission routes of Bartonella in populations of common vampire bats (Desmodus rotundus) in Peru and Belize, for which high infection prevalence has previously been reported. Phylogenetic analysis of the gltA gene for a subset of PCR-positive blood samples revealed sequences that were related to Bartonella described from vampire bats from Mexico, other Neotropical bat species, and streblid bat flies. Sequences associated with vampire bats clustered significantly by country but commonly spanned Central and South America, implying limited spatial structure. Stable and nonzero Bartonella prevalence between years supported endemic transmission in all sites. The odds of Bartonella infection for individual bats was unrelated to the intensity of bat flies ectoparasitism, but nearly all infected bats were infested, which precluded conclusive assessment of support for vector-borne transmission. While metagenomic sequencing found no strong evidence of Bartonella DNA in pooled bat saliva and fecal samples, we detected PCR positivity in individual saliva and feces, suggesting the potential for bacterial transmission through both direct contact (i.e., biting) and environmental (i.e., fecal) exposures. Further investigating the relative contributions of direct contact, environmental, and vector-borne transmission for bat Bartonella is an important next step to predict infection dynamics within bats and the risks of human and livestock exposures.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



References
Publication types
MeSH terms
Substances
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

