Protein Kinase A Signaling Inhibits Iridophore Differentiation in Zebrafish
- PMID: 30261583
- PMCID: PMC6315511
- DOI: 10.3390/jdb6040023
Protein Kinase A Signaling Inhibits Iridophore Differentiation in Zebrafish
Abstract
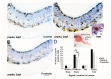
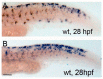
In zebrafish (Danio rerio), iridophores are specified from neural crest cells and represent a tractable system for examining mechanisms of cell fate and differentiation. Using this system, we have investigated the role of cAMP protein kinase A (PKA) signaling in pigment cell differentiation. Activation of PKA with the adenylyl cyclase activator forskolin reduces the number of differentiated iridophores in wildtype larvae, with insignificant changes to melanophore number. Inhibition of PKA with H89 significantly increases iridophore number, supporting a specific role for PKA during iridophore development. To determine the effects of altering PKA activity on iridophore and melanophore gene expression, we examined expression of iridophore marker pnp4a, melanophore marker mitfa, and the mitfa repressor foxd3. Consistent with our cell counts, forskolin significantly decreased pnp4a expression as detected by in situ hybridization and quantification of pnp4a+ cells. Forskolin had the opposite effect on mitfa and foxd3 gene activity, increasing the area of expression. As mitfa/nacre mutants have extra iridophores as compared to wildtype larvae, we examined the function of mitfa during PKA-sensitive iridophore development. Forskolin treatment of mitfa/nacre mutants did significantly reduce the number of iridophores but to a lesser extent than that observed in treated wildtype larvae. Taken together, our data suggests that PKA inhibits iridophore development in a subset of iridophore precursors, potentially via a foxd3-independent pathway.
Keywords: PKA signaling; differentiation; forskolin; iridophores; melanophores; mitfa; pnp4a.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

