Genome Wide Transcriptome Analysis Reveals Complex Regulatory Mechanisms Underlying Phosphate Homeostasis in Soybean Nodules
- PMID: 30261621
- PMCID: PMC6213598
- DOI: 10.3390/ijms19102924
Genome Wide Transcriptome Analysis Reveals Complex Regulatory Mechanisms Underlying Phosphate Homeostasis in Soybean Nodules
Abstract
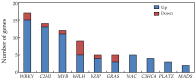
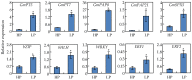
Phosphorus (P) deficiency is a major limitation for legume crop production. Although overall adaptations of plant roots to P deficiency have been extensively studied, only fragmentary information is available in regard to root nodule responses to P deficiency. In this study, genome wide transcriptome analysis was conducted using RNA-seq analysis in soybean nodules grown under P-sufficient (500 μM KH₂PO₄) and P-deficient (25 μM KH₂PO₄) conditions to investigate molecular mechanisms underlying soybean (Glycine max) nodule adaptation to phosphate (Pi) starvation. Phosphorus deficiency significantly decreased soybean nodule growth and nitrogenase activity. Nodule Pi concentrations declined by 49% in response to P deficiency, but this was well below the 87% and 88% decreases observed in shoots and roots, respectively. Nodule transcript profiling revealed that a total of 2055 genes exhibited differential expression patterns between Pi sufficient and deficient conditions. A set of (differentially expressed genes) DEGs appeared to be involved in maintaining Pi homeostasis in soybean nodules, including eight Pi transporters (PTs), eight genes coding proteins containing the SYG1/PHO81/XPR1 domain (SPXs), and 16 purple acid phosphatases (PAPs). The results suggest that a complex transcriptional regulatory network participates in soybean nodule adaption to Pi starvation, most notable a Pi signaling pathway, are involved in maintaining Pi homeostasis in nodules.
Keywords: P deficiency; RNA-seq; nodule; soybean; symbiotic nitrogen fixation.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

