Validation of Suitable Housekeeping Genes for the Normalization of mRNA Expression for Studying Tumor Acidosis
- PMID: 30261649
- PMCID: PMC6213411
- DOI: 10.3390/ijms19102930
Validation of Suitable Housekeeping Genes for the Normalization of mRNA Expression for Studying Tumor Acidosis
Abstract
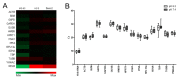
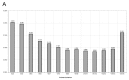
Similar to other types of cancer, acidification of tumor microenvironment is an important feature of osteosarcoma, and a major source of cellular stress that triggers cancer aggressiveness, drug resistance, and progression. Among the different effects of low extracellular pH on tumor cells, we have recently found that short-term exposure to acidosis strongly affects gene expression. This alteration might also occur for the most commonly used housekeeping genes (HKG), thereby causing erroneous interpretation of RT-qPCR data. On this basis, by using osteosarcoma cells cultured at different pH values, we aimed to identify the ideal HKG to be considered in studies on tumor-associated acidosis. We verified the stability of 15 commonly used HKG through five algorithms (NormFinder, geNorm, BestKeeper, ΔCT, coefficient of variation) and found that no universal HKG is suitable, since at least four HKG are necessary for proper normalization. Furthermore, according to the acceptable range of values, YWHAZ, GAPDH, GUSB, and 18S rRNA were the most stable reference genes at different pH. Our results will be helpful for future investigations focusing on the effect of altered microenvironment on cancer behavior, particularly on the effectiveness of anticancer therapies in acid conditions.
Keywords: RT-qPCR; acidosis; housekeeping gene; tumor microenvironment.
Conflict of interest statement
The authors declare no conflict of interest. The founding sponsors had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, and in the decision to publish the results.
Figures

Similar articles
-
Expression stability of common housekeeping genes is differently affected by bowel inflammation and cancer: implications for finding suitable normalizers for inflammatory bowel disease studies.Inflamm Bowel Dis. 2014 Jul;20(7):1147-56. doi: 10.1097/MIB.0000000000000067. Inflamm Bowel Dis. 2014. PMID: 24859296
-
Identification of the best housekeeping gene for RT-qPCR analysis of human pancreatic organoids.PLoS One. 2021 Dec 8;16(12):e0260902. doi: 10.1371/journal.pone.0260902. eCollection 2021. PLoS One. 2021. PMID: 34879096 Free PMC article.
-
Identification and Validation of Housekeeping Genes for Gene Expression Analysis of Cancer Stem Cells.PLoS One. 2016 Feb 19;11(2):e0149481. doi: 10.1371/journal.pone.0149481. eCollection 2016. PLoS One. 2016. PMID: 26894994 Free PMC article.
-
The dilution effect and the importance of selecting the right internal control genes for RT-qPCR: a paradigmatic approach in fetal sheep.BMC Res Notes. 2015 Feb 27;8:58. doi: 10.1186/s13104-015-0973-7. BMC Res Notes. 2015. PMID: 25881111 Free PMC article.
-
Validation of Housekeeping Genes for Gene Expression Profiling in Fish: A Necessity.Crit Rev Eukaryot Gene Expr. 2019;29(6):565-579. doi: 10.1615/CritRevEukaryotGeneExpr.2019028964. Crit Rev Eukaryot Gene Expr. 2019. PMID: 32422011 Review.
Cited by
-
Housekeeping Gene Expression in the Fetal and Neonatal Murine Thymus Following Coxsackievirus B4 Infection.Genes (Basel). 2020 Mar 5;11(3):279. doi: 10.3390/genes11030279. Genes (Basel). 2020. PMID: 32150956 Free PMC article.
-
Acid-Induced Inflammatory Cytokines in Osteoblasts: A Guided Path to Osteolysis in Bone Metastasis.Front Cell Dev Biol. 2021 May 28;9:678532. doi: 10.3389/fcell.2021.678532. eCollection 2021. Front Cell Dev Biol. 2021. PMID: 34124067 Free PMC article.
-
Real-time PCR quantification of spliced X-box binding protein 1 (XBP1) using a universal primer method.PLoS One. 2019 Jul 22;14(7):e0219978. doi: 10.1371/journal.pone.0219978. eCollection 2019. PLoS One. 2019. PMID: 31329612 Free PMC article.
-
Housekeeping gene expression variability in differentiating and non-differentiating 3T3-L1 cells.Adipocyte. 2023 Dec;12(1):2235081. doi: 10.1080/21623945.2023.2235081. Adipocyte. 2023. PMID: 37436361 Free PMC article.
-
Innovative GenExpA software for selecting suitable reference genes for reliable normalization of gene expression in melanoma.Sci Rep. 2022 Feb 28;12(1):3331. doi: 10.1038/s41598-022-07257-6. Sci Rep. 2022. PMID: 35228606 Free PMC article.
References
-
- Warburg O. On the origin of cancer cells. Science. 1956;123:309–314. - PubMed
-
- Lemma S., Di Pompo G., Porporato P.E., Sboarina M., Russell S., Gillies R.J., Baldini N., Sonveaux P., Avnet S. MDA-MB-231 breast cancer cells fuel osteoclast metabolism and activity: A new rationale for the pathogenesis of osteolytic bone metastases. Biochim. Biophys. Acta. 2017;1863:3254–3264. doi: 10.1016/j.bbadis.2017.08.030. - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

