Developmental enhancers and chromosome topology
- PMID: 30262496
- PMCID: PMC6986801
- DOI: 10.1126/science.aau0320
Developmental enhancers and chromosome topology
Abstract
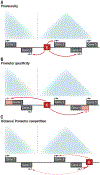
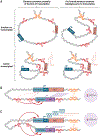
Developmental enhancers mediate on/off patterns of gene expression in specific cell types at particular stages during metazoan embryogenesis. They typically integrate multiple signals and regulatory determinants to achieve precise spatiotemporal expression. Such enhancers can map quite far-one megabase or more-from the genes they regulate. How remote enhancers relay regulatory information to their target promoters is one of the central mysteries of genome organization and function. A variety of contrasting mechanisms have been proposed over the years, including enhancer tracking, linking, looping, and mobilization to transcription factories. We argue that extreme versions of these mechanisms cannot account for the transcriptional dynamics and precision seen in living cells, tissues, and embryos. We describe emerging evidence for dynamic three-dimensional hubs that combine different elements of the classical models.
Copyright © 2018, American Association for the Advancement of Science.
Conflict of interest statement
Figures
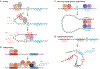
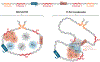
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

