Clinical cancer genomic profiling by three-platform sequencing of whole genome, whole exome and transcriptome
- PMID: 30262806
- PMCID: PMC6160438
- DOI: 10.1038/s41467-018-06485-7
Clinical cancer genomic profiling by three-platform sequencing of whole genome, whole exome and transcriptome
Abstract
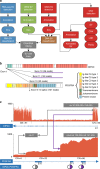
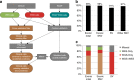
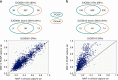
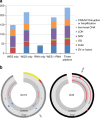
To evaluate the potential of an integrated clinical test to detect diverse classes of somatic and germline mutations relevant to pediatric oncology, we performed three-platform whole-genome (WGS), whole exome (WES) and transcriptome (RNA-Seq) sequencing of tumors and normal tissue from 78 pediatric cancer patients in a CLIA-certified, CAP-accredited laboratory. Our analysis pipeline achieves high accuracy by cross-validating variants between sequencing types, thereby removing the need for confirmatory testing, and facilitates comprehensive reporting in a clinically-relevant timeframe. Three-platform sequencing has a positive predictive value of 97-99, 99, and 91% for somatic SNVs, indels and structural variations, respectively, based on independent experimental verification of 15,225 variants. We report 240 pathogenic variants across all cases, including 84 of 86 known from previous diagnostic testing (98% sensitivity). Combined WES and RNA-Seq, the current standard for precision oncology, achieved only 78% sensitivity. These results emphasize the critical need for incorporating WGS in pediatric oncology testing.
Conflict of interest statement
The authors declare no competing interests.
Figures

Similar articles
-
Inconsistency and features of single nucleotide variants detected in whole exome sequencing versus transcriptome sequencing: A case study in lung cancer.Methods. 2015 Jul 15;83:118-27. doi: 10.1016/j.ymeth.2015.04.016. Epub 2015 Apr 23. Methods. 2015. PMID: 25913717 Free PMC article.
-
Whole-genome sequencing is more powerful than whole-exome sequencing for detecting exome variants.Proc Natl Acad Sci U S A. 2015 Apr 28;112(17):5473-8. doi: 10.1073/pnas.1418631112. Epub 2015 Mar 31. Proc Natl Acad Sci U S A. 2015. PMID: 25827230 Free PMC article.
-
Analytical validation of whole exome and whole genome sequencing for clinical applications.BMC Med Genomics. 2014 Apr 23;7:20. doi: 10.1186/1755-8794-7-20. BMC Med Genomics. 2014. PMID: 24758382 Free PMC article.
-
Exome versus transcriptome sequencing in identifying coding region variants.Expert Rev Mol Diagn. 2012 Apr;12(3):241-51. doi: 10.1586/erm.12.10. Expert Rev Mol Diagn. 2012. PMID: 22468815 Review.
-
Use of whole exome and genome sequencing in the identification of genetic causes of primary immunodeficiencies.Curr Opin Allergy Clin Immunol. 2012 Dec;12(6):623-8. doi: 10.1097/ACI.0b013e3283588ca6. Curr Opin Allergy Clin Immunol. 2012. PMID: 23095910 Review.
Cited by
-
Tumor Variant Identification That Accounts for the Unique Molecular Landscape of Pediatric Malignancies.JNCI Cancer Spectr. 2018 Oct;2(4):pky079. doi: 10.1093/jncics/pky079. Epub 2019 Jan 25. JNCI Cancer Spectr. 2018. PMID: 30976750 Free PMC article.
-
Etiology of oncogenic fusions in 5,190 childhood cancers and its clinical and therapeutic implication.Nat Commun. 2023 Apr 5;14(1):1739. doi: 10.1038/s41467-023-37438-4. Nat Commun. 2023. PMID: 37019972 Free PMC article.
-
Sequential filtering for clinically relevant variants as a method for clinical interpretation of whole exome sequencing findings in glioma.BMC Med Genomics. 2021 Feb 23;14(1):54. doi: 10.1186/s12920-021-00904-3. BMC Med Genomics. 2021. PMID: 33622343 Free PMC article.
-
Genomic profiling of circulating tumor DNA for childhood cancers.Leukemia. 2025 Feb;39(2):420-430. doi: 10.1038/s41375-024-02461-x. Epub 2024 Nov 10. Leukemia. 2025. PMID: 39523434
-
Pan-cancer image-based detection of clinically actionable genetic alterations.Nat Cancer. 2020 Aug;1(8):789-799. doi: 10.1038/s43018-020-0087-6. Epub 2020 Jul 27. Nat Cancer. 2020. PMID: 33763651 Free PMC article.
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

