Specificity of the osmotic stress response in Candida albicans highlighted by quantitative proteomics
- PMID: 30262823
- PMCID: PMC6160413
- DOI: 10.1038/s41598-018-32792-6
Specificity of the osmotic stress response in Candida albicans highlighted by quantitative proteomics
Abstract
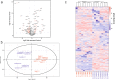
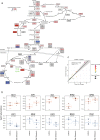
Stress adaptation is critical for the survival of microbes in dynamic environments, and in particular, for fungal pathogens to survive in and colonise host niches. Proteomic analyses have the potential to significantly enhance our understanding of these adaptive responses by providing insight into post-transcriptional regulatory mechanisms that contribute to the outputs, as well as testing presumptions about the regulation of protein levels based on transcript profiling. Here, we used label-free, quantitative mass spectrometry to re-examine the response of the major fungal pathogen of humans, Candida albicans, to osmotic stress. Of the 1,262 proteins that were identified, 84 were down-regulated in response to 1M NaCl, reflecting the decrease in ribosome biogenesis and translation that often accompanies stress. The 64 up-regulated proteins included central metabolic enzymes required for glycerol synthesis, a key osmolyte for this yeast, as well as proteins with functions during stress. These data reinforce the view that adaptation to salt stress involves a transient reduction in ribosome biogenesis and translation together with the accumulation of the osmolyte, glycerol. The specificity of the response to salt stress is highlighted by the small proportion of quantified C. albicans proteins (5%) whose relative elevated abundances were statistically significant.
Conflict of interest statement
L.A.G., J.I.L. and J.P.C.V. are employed by Waters Corporation, which operates in the field covered by the article. The remaining authors declare no competing financial interests.
Figures




Similar articles
-
Metabolic Reprogramming in the Opportunistic Yeast Candida albicans in Response to Hypoxia.mSphere. 2020 Feb 26;5(1):e00913-19. doi: 10.1128/mSphere.00913-19. mSphere. 2020. PMID: 32102943 Free PMC article.
-
Potential role of lysine acetylation in the stepwise adaptation of Candida albicans to fluconazole.Microbiol Spectr. 2025 May 6;13(5):e0279724. doi: 10.1128/spectrum.02797-24. Epub 2025 Apr 15. Microbiol Spectr. 2025. PMID: 40231831 Free PMC article.
-
Candida albicans response regulator gene SSK1 regulates a subset of genes whose functions are associated with cell wall biosynthesis and adaptation to oxidative stress.Eukaryot Cell. 2003 Oct;2(5):1018-24. doi: 10.1128/EC.2.5.1018-1024.2003. Eukaryot Cell. 2003. PMID: 14555484 Free PMC article.
-
From commensal to pathogen: stage- and tissue-specific gene expression of Candida albicans.Curr Opin Microbiol. 2004 Aug;7(4):336-41. doi: 10.1016/j.mib.2004.06.003. Curr Opin Microbiol. 2004. PMID: 15288621 Review.
-
Stress adaptation in a pathogenic fungus.J Exp Biol. 2014 Jan 1;217(Pt 1):144-55. doi: 10.1242/jeb.088930. J Exp Biol. 2014. PMID: 24353214 Free PMC article. Review.
Cited by
-
Microbial Biocontrol Agents and Natural Products Act as Salt Stress Mitigators in Lactuca sativa L.Plants (Basel). 2024 Sep 6;13(17):2505. doi: 10.3390/plants13172505. Plants (Basel). 2024. PMID: 39273989 Free PMC article.
-
Metabolic Plasticity of Candida albicans in Response to Different Environmental Conditions.J Fungi (Basel). 2022 Jul 12;8(7):723. doi: 10.3390/jof8070723. J Fungi (Basel). 2022. PMID: 35887478 Free PMC article.
-
Comparative Genomics and Transcriptomics Analyses Reveal a Unique Environmental Adaptability of Vibrio fujianensis.Microorganisms. 2020 Apr 13;8(4):555. doi: 10.3390/microorganisms8040555. Microorganisms. 2020. PMID: 32294952 Free PMC article.
-
SPT20 Regulates the Hog1-MAPK Pathway and Is Involved in Candida albicans Response to Hyperosmotic Stress.Front Microbiol. 2020 Feb 21;11:213. doi: 10.3389/fmicb.2020.00213. eCollection 2020. Front Microbiol. 2020. PMID: 32153525 Free PMC article.
-
Deciphering cargo contents in extracellular vesicles of Candida haemulonii var. vulnera.Comput Struct Biotechnol J. 2025 Apr 15;27:1887-1900. doi: 10.1016/j.csbj.2025.04.014. eCollection 2025. Comput Struct Biotechnol J. 2025. PMID: 40487199 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
- BB/K017365/1/Biotechnology and Biological Sciences Research Council (BBSRC)/International
- BB/F005210/1/Biotechnology and Biological Sciences Research Council (BBSRC)/International
- 097377/WT_/Wellcome Trust/United Kingdom
- BB/F005210/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- MR/M026663/1/MRC_/Medical Research Council/United Kingdom
- BB/F00513X/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- MR/N006364/1/MRC_/Medical Research Council/United Kingdom
- BB/F00513X/1/Biotechnology and Biological Sciences Research Council (BBSRC)/International
- MR/M026663/1/Medical Research Council (MRC)/International
- BB/C007433/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

