The soybean Rhg1 amino acid transporter gene alters glutamate homeostasis and jasmonic acid-induced resistance to soybean cyst nematode
- PMID: 30264924
- PMCID: PMC6637870
- DOI: 10.1111/mpp.12753
The soybean Rhg1 amino acid transporter gene alters glutamate homeostasis and jasmonic acid-induced resistance to soybean cyst nematode
Abstract
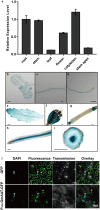
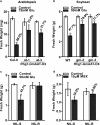
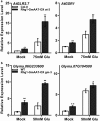
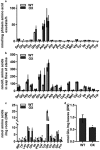
Rhg1 (resistance to Heterodera glycines 1) is an important locus that contributes to resistance against soybean cyst nematode (SCN; Heterodera glycines Ichinohe), which is the most economically damaging disease of soybean worldwide. Simultaneous overexpression of three genes encoding a predicted amino acid transporter, an α-soluble N-ethylmaleimide-sensitive factor attachment protein (α-SNAP) and a predicted wound-induced protein resulted in resistance to SCN provided by this locus. However, the roles of two of these genes (excluding α-SNAP) remain unknown. Here, we report the functional characterization of Glyma.18G022400, a gene at the Rhg1 locus that encodes the predicted amino acid transporter Rhg1-GmAAT. Although the direct role of Rhg1-GmAAT in glutamate transport was not demonstrated, multiple lines of evidence showed that Rhg1-GmAAT impacts glutamic acid tolerance and glutamate transportation in soybean. Transcriptomic and metabolite profiling indicated that overexpression of Rhg1-GmAAT activated the jasmonic acid (JA) pathway. Treatment with a JA biosynthesis inhibitor reduced the resistance provided by the Rhg1-containing PI88788 to SCN, which suggested that the JA pathway might play a role in Rhg1-mediated resistance to SCN. Our results could be helpful for the clarification of the mechanism of resistance to SCN provided by Rhg1 in soybean.
Keywords: Rhg1; amino acid transporter; glutamate; jasmonic acid; soybean; soybean cyst nematode.
© 2018 The Authors. Molecular Plant Pathology Published by BSPP and John Wiley & Sons Ltd.
Figures


Similar articles
-
t-SNAREs bind the Rhg1 α-SNAP and mediate soybean cyst nematode resistance.Plant J. 2020 Oct;104(2):318-331. doi: 10.1111/tpj.14923. Epub 2020 Aug 6. Plant J. 2020. PMID: 32645235
-
Soybean transporter AAT Rhg1 abundance increases along the nematode migration path and impacts vesiculation and ROS.Plant Physiol. 2023 May 2;192(1):133-153. doi: 10.1093/plphys/kiad098. Plant Physiol. 2023. PMID: 36805759 Free PMC article.
-
Disease resistance through impairment of α-SNAP-NSF interaction and vesicular trafficking by soybean Rhg1.Proc Natl Acad Sci U S A. 2016 Nov 22;113(47):E7375-E7382. doi: 10.1073/pnas.1610150113. Epub 2016 Nov 7. Proc Natl Acad Sci U S A. 2016. PMID: 27821740 Free PMC article.
-
Exploring Soybean Resistance to Soybean Cyst Nematode.Annu Rev Phytopathol. 2022 Aug 26;60:379-409. doi: 10.1146/annurev-phyto-020620-120823. Epub 2022 May 19. Annu Rev Phytopathol. 2022. PMID: 35587510 Review.
-
Soybean Resistance to the Soybean Cyst Nematode Heterodera glycines: An Update.Phytopathology. 2016 Dec;106(12):1444-1450. doi: 10.1094/PHYTO-06-16-0227-RVW. Epub 2016 Sep 6. Phytopathology. 2016. PMID: 27392178 Review.
Cited by
-
Genome-Wide Identification of Rapid Alkalinization Factor Family in Brassica napus and Functional Analysis of BnRALF10 in Immunity to Sclerotinia sclerotiorum.Front Plant Sci. 2022 May 3;13:877404. doi: 10.3389/fpls.2022.877404. eCollection 2022. Front Plant Sci. 2022. PMID: 35592581 Free PMC article.
-
TaWI12 may be involved in pistillody and leaf cracking in wheat.BMC Plant Biol. 2025 Jan 29;25(1):123. doi: 10.1186/s12870-025-06133-5. BMC Plant Biol. 2025. PMID: 39875850 Free PMC article.
-
Molecular and Cellular Mechanisms Involved in Host-Specific Resistance to Cyst Nematodes in Crops.Front Plant Sci. 2021 Mar 9;12:641582. doi: 10.3389/fpls.2021.641582. eCollection 2021. Front Plant Sci. 2021. PMID: 33767723 Free PMC article. Review.
-
Amino acid transporter gene TaATLa1 from Triticum aestivum L. improves growth under nitrogen sufficiency and is down regulated under nitrogen deficiency.Planta. 2022 Aug 29;256(4):65. doi: 10.1007/s00425-022-03978-0. Planta. 2022. PMID: 36036331
-
Biocontrol Potential of Bacillus strains against soybean cyst nematode (Heterodera glycines) and for promotion of soybean growth.BMC Microbiol. 2024 Sep 28;24(1):371. doi: 10.1186/s12866-024-03514-y. BMC Microbiol. 2024. PMID: 39342079 Free PMC article.
References
-
- Arelli, A.P. , Wilcox, J.A. , Myers, O. and Gibson, P.T. (1997) Soybean germplasm resistant to races 1 and 2 of Heterodera glycines . Crop Sci. 37, 1367–1369.
-
- Avanci, N.C. , Luche, D.D. , Goldman, G.H. and Goldman, M.H. (2010) Jasmonates are phytohormones with multiple functions, including plant defense and reproduction. Genet. Mol. Res. 9, 484–505. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

