Dynamic epimarks in sex-related genes predict gonad phenotype in the European sea bass, a fish with mixed genetic and environmental sex determination
- PMID: 30265213
- PMCID: PMC6284782
- DOI: 10.1080/15592294.2018.1529504
Dynamic epimarks in sex-related genes predict gonad phenotype in the European sea bass, a fish with mixed genetic and environmental sex determination
Abstract
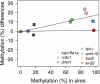
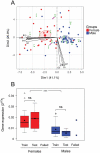
The integration of genomic and environmental influences into methylation patterns to bring about a phenotype is of central interest in developmental epigenetics, but many details are still unclear. The sex ratios of the species used here, the European sea bass, are determined by genetic and temperature influences. We created four families from parents known to produce offspring with different sex ratios, exposed larvae to masculinizing temperatures and examined, in juvenile gonads, the DNA methylation of seven genes related to sexual development by a targeted sequencing approach. The genes most affected by both genetics and environment were cyp19a1a and dmrt1, with contrasting sex-specific methylation and temperature responses. The relationship between cyp19a1a methylation and expression is relevant to the epigenetic regulation of vertebrate sex, and we report the evidence of such relationship only below a methylation threshold, ~ 80%, and that it was sex-specific: negatively correlated in females but positively correlated in males. From parents to offspring, the methylation in gonads was midway between oocytes and sperm, with bias towards oocytes for amh-r2, er-β2, fsh-r and cyp19a1a. In contrast, dmrt1 levels resembled those of sperm. The methylation of individual CpGs from foxl2, er-β2 and nr3c1 were conserved from parents to offspring, whereas those of cyp19a1a, dmrt1 and amh-r2 were affected by temperature. Utilizing a machine-learning procedure based on the methylation levels of a selected set of CpGs, we present the first, to our knowledge, system based on epigenetic marks capable of predicting sex in an animal with ~ 90% accuracy and discuss possible applications.
Keywords: DNA methylation; DNA methylation threshold; Early development; aromatase; cyp19a1a; dmrt1; environmental temperature; epigenetic inheritance; epigenetic marks; sex determination.
Figures




References
-
- Gilbert SF, Epel D.. Ecological developmental biology: integrating epigenetics, medicine, and evolution. Sunderland, MA: Sinauer Associates; 2008.
-
- Piferrer F. Epigenetics of sex determination and gonadogenesis. Dev Dyn. 2013;242:360–370. - PubMed
-
- Devlin RH, Nagahama Y. Sex determination and sex differentiation in fish: an overview of genetic, physiological, and environmental influences. Aquaculture. 2002;208:191–364.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
