Identification, Localization, and Quantification of HIV Reservoirs Using Microscopy
- PMID: 30265439
- PMCID: PMC6386609
- DOI: 10.1002/cpcb.64
Identification, Localization, and Quantification of HIV Reservoirs Using Microscopy
Abstract
The major barrier to eradicating human immunodeficiency virus-1 (HIV) infection is the generation and extended survival of HIV reservoirs. In order to eradicate HIV infection, it is essential to detect, quantify, and characterize circulating and tissue-associated viral reservoirs in infected individuals. Currently, PCR-based technologies and Quantitative Viral Outgrowth Assays (Q-VOA) are the gold standards to detect viral reservoirs. However, these methods are limited to detecting circulating viral reservoirs, and it has been shown that they misrepresent the size of the reservoirs, largely because they detect only one component of the HIV life cycle and are unable to detect viral reservoirs in tissues. Here, we described the use of multiple detection systems to identify integrated HIV DNA or viral mRNA and several HIV proteins in circulating and tissue reservoirs using improved staining and microscopy techniques. We believe that this imaging-based approach for detecting HIV reservoirs will lead to breakthroughs necessary to eradicate these reservoirs. © 2018 by John Wiley & Sons, Inc.
Keywords: AIDS; Q-VOA; T cells; astrocytes; macrophages; transmission.
© 2018 John Wiley & Sons, Inc.
Conflict of interest statement
Figures
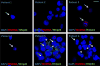
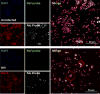

Similar articles
-
Ultrasensitive HIV-1 p24 Assay Detects Single Infected Cells and Differences in Reservoir Induction by Latency Reversal Agents.J Virol. 2017 Feb 28;91(6):e02296-16. doi: 10.1128/JVI.02296-16. Print 2017 Mar 15. J Virol. 2017. PMID: 28077644 Free PMC article.
-
Identification, Quantification, and Characterization of HIV-1 Reservoirs in the Human Brain.Cells. 2022 Aug 2;11(15):2379. doi: 10.3390/cells11152379. Cells. 2022. PMID: 35954221 Free PMC article.
-
Single-cell characterization and quantification of translation-competent viral reservoirs in treated and untreated HIV infection.PLoS Pathog. 2019 Feb 27;15(2):e1007619. doi: 10.1371/journal.ppat.1007619. eCollection 2019 Feb. PLoS Pathog. 2019. PMID: 30811499 Free PMC article.
-
Combating the HIV reservoirs.Biotechnol Genet Eng Rev. 2018 Apr;34(1):76-89. doi: 10.1080/02648725.2018.1471641. Epub 2018 May 21. Biotechnol Genet Eng Rev. 2018. PMID: 29781356 Review.
-
How to best measure HIV reservoirs?Curr Opin HIV AIDS. 2013 May;8(3):170-5. doi: 10.1097/COH.0b013e32835fc619. Curr Opin HIV AIDS. 2013. PMID: 23564004 Free PMC article. Review.
Cited by
-
HIV infection and latency induce a unique metabolic signature in human macrophages.Sci Rep. 2019 Mar 8;9(1):3941. doi: 10.1038/s41598-019-39898-5. Sci Rep. 2019. PMID: 30850623 Free PMC article.
-
HIV-1 reservoirs in urethral macrophages of patients under suppressive antiretroviral therapy.Nat Microbiol. 2019 Apr;4(4):633-644. doi: 10.1038/s41564-018-0335-z. Epub 2019 Feb 4. Nat Microbiol. 2019. PMID: 30718846
-
Astrocytes are HIV reservoirs in the brain: A cell type with poor HIV infectivity and replication but efficient cell-to-cell viral transfer.J Neurochem. 2021 Jul;158(2):429-443. doi: 10.1111/jnc.15336. Epub 2021 Mar 22. J Neurochem. 2021. PMID: 33655498 Free PMC article.
-
The Landscape of IFN/ISG Signaling in HIV-1-Infected Macrophages and Its Possible Role in the HIV-1 Latency.Cells. 2021 Sep 9;10(9):2378. doi: 10.3390/cells10092378. Cells. 2021. PMID: 34572027 Free PMC article. Review.
-
The hypoxia-regulated ectonucleotidase CD73 is a host determinant of HIV latency.Cell Rep. 2023 Nov 28;42(11):113285. doi: 10.1016/j.celrep.2023.113285. Epub 2023 Oct 31. Cell Rep. 2023. PMID: 37910505 Free PMC article.
References
-
- Banga R, Procopio FA, Perreau M. Current approaches to assess HIV-1 persistence. Curr Opin HIV AIDS. 2016;11(4):424–431. - PubMed
-
- Blankson J, Persaud D, Siliciano RF. Latent reservoirs for HIV-1. Curr Opin Infect Dis. 1999;12(1):5–11. - PubMed
-
- Bussolati G, Gugliotta P, Volante M, Pace M, Papotti M. Retrieved endogenous biotin: a novel marker and a potential pitfall in diagnostic immunohistochemistry. Histopathology. 1997;31(5):400–407. - PubMed
-
- Churchill MJ, Deeks SG, Margolis DM, Siliciano RF, Swanstrom R. HIV reservoirs: what, where and how to target them. Nat Rev Microbiol. 2016;14(1):55–60. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

