Split inactivated COBRA vaccine elicits protective antibodies against H1N1 and H3N2 influenza viruses
- PMID: 30265682
- PMCID: PMC6161894
- DOI: 10.1371/journal.pone.0204284
Split inactivated COBRA vaccine elicits protective antibodies against H1N1 and H3N2 influenza viruses
Erratum in
-
Correction: Split inactivated COBRA vaccine elicits protective antibodies against H1N1 and H3N2 influenza viruses.PLoS One. 2018 Dec 27;13(12):e0210043. doi: 10.1371/journal.pone.0210043. eCollection 2018. PLoS One. 2018. PMID: 30589902 Free PMC article.
Expression of concern in
-
Expression of Concern: Split inactivated COBRA vaccine elicits protective antibodies against H1N1 and H3N2 influenza viruses.PLoS One. 2024 Jan 10;19(1):e0297253. doi: 10.1371/journal.pone.0297253. eCollection 2024. PLoS One. 2024. PMID: 38198500 Free PMC article. No abstract available.
Abstract
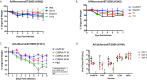
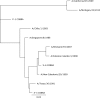
Development of broadly reactive or universal influenza vaccines will be a paradigm shifting event for the influenza vaccine field. These next generation vaccines could replace the current standard of care with vaccines that elicit broadly cross-protective immune responses. However, a variety of in vitro and in vivo models are necessary to make the best assessments of these vaccine formulations to determine their mechanisms of action, and allow for downselection of candidates prior to human clinical trials. Our group has developed the computationally optimized broadly reactive antigen (COBRA) technology to develop HA head-based strategies to elicit antibodies against H1, H3, and H5 influenza strains. These vaccines elicit broadly reactive antibody responses that neutralize not only historical and contemporary vaccine strains, but also co-circulating variants in mice. In this study, we used H1 and H3 HA antigens in a split, inactivated vaccine (IIV) formulation in combination with the AF03 squalene-in-water emulsion adjuvant in ferrets immunologically naïve to influenza virus. The H3 COBRA IIV vaccine T11 elicited antibodies with HAI activity against more H3N2 influenza strains compared to IIV expressing wild-type H3 HA antigens, except for IIV vaccines expressing the HA from A/Texas/50/2012 (Tx/12) virus. H1 COBRA IIV vaccines, P1 and X6, elicited antibodies that recognized a similar number of H1N1 viruses as those antibodies elicited by IIV expressing the A/California/07/2009 (CA/09) HA. Ferrets vaccinated with the P1 or X6 COBRA IIV were protected against CA/09 challege and cleared virus from the lungs of the ferrets, similar to ferrets vaccinated with the CA/09 IIV.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
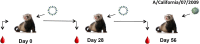



References
-
- Geeraedts F, Goutagny N, Hornung V, Severa M, de Haan A, Pool J, et al. Superior immunogenicity of inactivated whole virus H5N1 influenza vaccine is primarily controlled by Toll-like receptor signalling. PLoS Pathog. 2008;4(8):e1000138 10.1371/journal.ppat.1000138 ; PubMed Central PMCID: PMCPMC2516931. - DOI - PMC - PubMed
-
- Geeraedts F, Bungener L, Pool J, ter Veer W, Wilschut J, Huckriede A. Whole inactivated virus influenza vaccine is superior to subunit vaccine in inducing immune responses and secretion of proinflammatory cytokines by DCs. Influenza Other Respir Viruses. 2008;2(2):41–51. 10.1111/j.1750-2659.2008.00038.x ; PubMed Central PMCID: PMCPMC4941893. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

