RNA-Binding Protein HuR Regulates Both Mutant and Wild-Type IDH1 in IDH1-Mutated Cancer
- PMID: 30266754
- PMCID: PMC6359963
- DOI: 10.1158/1541-7786.MCR-18-0557
RNA-Binding Protein HuR Regulates Both Mutant and Wild-Type IDH1 in IDH1-Mutated Cancer
Abstract
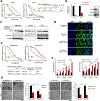
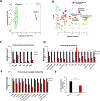
Isocitrate dehydrogenase 1 (IDH1) is the most commonly mutated metabolic enzyme in human malignancy. A heterozygous genetic alteration, arginine 132, promotes the conversion of α-ketoglutarate to D-2-hydroxyglutarate (2-HG). Although pharmacologic inhibitors of mutant IDH1 are promising, resistance mechanisms to targeted therapy are not understood. Additionally, the role of wild-type IDH1 (WT.IDH1) in cancer requires further study. Recently, it was observed that the regulatory RNA-binding protein, HuR (ELAVL1), protects nutrient-deprived cancer cells without IDH1 mutations, by stabilizing WT.IDH1 transcripts. In the present study, a similar regulatory effect on both mutant (Mut.IDH1) and WT.IDH1 transcripts in heterozygous IDH1-mutant tumors is observed. In ribonucleoprotein immunoprecipitation assays of IDH1-mutant cell lines, wild-type and mutant IDH1 mRNAs each bound to HuR. Both isoforms were profoundly downregulated at the mRNA and protein levels after genetic suppression of HuR (siRNAs or CRISPR deletion) in HT1080 (R132C IDH1 mutation) and BT054 cells (R132H). Proliferation and invasion were adversely affected after HuR suppression and metabolomic studies revealed a reduction in Pentose Phosphate Pathway metabolites, nucleotide precursors, and 2-HG levels. HuR-deficient cells were especially sensitive to stress, including low glucose conditions or a mutant IDH1 inhibitor (AGI-5198). IDH1-mutant cancer cells were rescued by WT.IDH1 overexpression to a greater extent than Mut.IDH1 overexpression under these conditions. This study reveals the importance of HuR's regulation of both mutant and wild-type IDH1 in tumors harboring a heterozygous IDH1 mutation with implications for therapy. IMPLICATIONS: This study highlights the HuR-IDH1 (mutant and wild-type IDH1) regulatory axis as a critical, actionable therapeutic target in IDH1-mutated cancer, and incomplete blockade of the entire HuR-IDH1 survival axis would likely diminish the efficacy of drugs that selectively target only the mutant isoenzyme.
©2018 American Association for Cancer Research.
Conflict of interest statement
Figures





Similar articles
-
Posttranscriptional Upregulation of IDH1 by HuR Establishes a Powerful Survival Phenotype in Pancreatic Cancer Cells.Cancer Res. 2017 Aug 15;77(16):4460-4471. doi: 10.1158/0008-5472.CAN-17-0015. Epub 2017 Jun 26. Cancer Res. 2017. PMID: 28652247 Free PMC article.
-
Mutant IDH1 is required for IDH1 mutated tumor cell growth.Oncotarget. 2012 Aug;3(8):774-82. doi: 10.18632/oncotarget.577. Oncotarget. 2012. PMID: 22885298 Free PMC article.
-
Synthesis and evaluation of radiolabeled AGI-5198 analogues as candidate radiotracers for imaging mutant IDH1 expression in tumors.Bioorg Med Chem Lett. 2018 Feb 15;28(4):694-699. doi: 10.1016/j.bmcl.2018.01.015. Epub 2018 Jan 12. Bioorg Med Chem Lett. 2018. PMID: 29366652 Free PMC article.
-
Wild-type and mutated IDH1/2 enzymes and therapy responses.Oncogene. 2018 Apr;37(15):1949-1960. doi: 10.1038/s41388-017-0077-z. Epub 2018 Jan 25. Oncogene. 2018. PMID: 29367755 Free PMC article. Review.
-
Understanding and targeting the disease-related RNA binding protein human antigen R (HuR).Wiley Interdiscip Rev RNA. 2020 May;11(3):e1581. doi: 10.1002/wrna.1581. Epub 2020 Jan 23. Wiley Interdiscip Rev RNA. 2020. PMID: 31970930 Free PMC article. Review.
Cited by
-
ELAV/Hu RNA-binding protein family: key regulators in neurological disorders, cancer, and other diseases.RNA Biol. 2025 Dec;22(1):1-11. doi: 10.1080/15476286.2025.2471133. Epub 2025 Mar 20. RNA Biol. 2025. PMID: 40000387 Free PMC article. Review.
-
Limited nutrient availability in the tumor microenvironment renders pancreatic tumors sensitive to allosteric IDH1 inhibitors.Nat Cancer. 2022 Jul;3(7):852-865. doi: 10.1038/s43018-022-00393-y. Epub 2022 Jun 9. Nat Cancer. 2022. PMID: 35681100 Free PMC article.
-
Increased glucose availability sensitizes pancreatic cancer to chemotherapy.Nat Commun. 2023 Jun 28;14(1):3823. doi: 10.1038/s41467-023-38921-8. Nat Commun. 2023. PMID: 37380658 Free PMC article.
-
The regulatory mechanisms and inhibitors of isocitrate dehydrogenase 1 in cancer.Acta Pharm Sin B. 2023 Apr;13(4):1438-1466. doi: 10.1016/j.apsb.2022.12.019. Epub 2023 Feb 2. Acta Pharm Sin B. 2023. PMID: 37139412 Free PMC article. Review.
-
IDH1 Inhibition Potentiates Chemotherapy Efficacy in Pancreatic Cancer.Cancer Res. 2024 Sep 16;84(18):3072-3085. doi: 10.1158/0008-5472.CAN-23-1895. Cancer Res. 2024. PMID: 38843355 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

