N-Deacetylases required for muramic-δ-lactam production are involved in Clostridium difficile sporulation, germination, and heat resistance
- PMID: 30266804
- PMCID: PMC6254358
- DOI: 10.1074/jbc.RA118.004273
N-Deacetylases required for muramic-δ-lactam production are involved in Clostridium difficile sporulation, germination, and heat resistance
Abstract
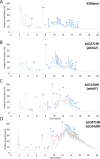
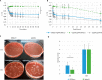
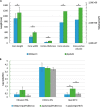
Spores are produced by many organisms as a survival mechanism activated in response to several environmental stresses. Bacterial spores are multilayered structures, one of which is a peptidoglycan layer called the cortex, containing muramic-δ-lactams that are synthesized by at least two bacterial enzymes, the muramoyl-l-alanine amidase CwlD and the N-deacetylase PdaA. This study focused on the spore cortex of Clostridium difficile, a Gram-positive, toxin-producing anaerobic bacterial pathogen that can colonize the human intestinal tract and is a leading cause of antibiotic-associated diarrhea. Using ultra-HPLC coupled with high-resolution MS, here we found that the spore cortex of the C. difficile 630Δerm strain differs from that of Bacillus subtilis Among these differences, the muramic-δ-lactams represented only 24% in C. difficile, compared with 50% in B. subtilis CD630_14300 and CD630_27190 were identified as genes encoding the C. difficile N-deacetylases PdaA1 and PdaA2, required for muramic-δ-lactam synthesis. In a pdaA1 mutant, only 0.4% of all muropeptides carried a muramic-δ-lactam modification, and muramic-δ-lactams were absent in the cortex of a pdaA1-pdaA2 double mutant. Of note, the pdaA1 mutant exhibited decreased sporulation, altered germination, decreased heat resistance, and delayed virulence in a hamster infection model. These results suggest a much greater role for muramic-δ-lactams in C. difficile than in other bacteria, including B. subtilis In summary, the spore cortex of C. difficile contains lower levels of muramic-δ-lactams than that of B. subtilis, and PdaA1 is the major N-deacetylase for muramic-δ-lactam biosynthesis in C. difficile, contributing to sporulation, heat resistance, and virulence.
Keywords: Clostridium difficile; Gram-positive bacteria; N-deacetylase; acetylation; bacteria; bacterial pathogenesis; germination; infectious disease; microbiology; muramic-δ-lactams; peptidoglycan; spore cortex; sporulation.
© 2018 Coullon et al.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article
Figures



Similar articles
-
Production of muramic delta-lactam in Bacillus subtilis spore peptidoglycan.J Bacteriol. 2004 Jan;186(1):80-9. doi: 10.1128/JB.186.1.80-89.2004. J Bacteriol. 2004. PMID: 14679227 Free PMC article.
-
Clostridium difficile Lipoprotein GerS Is Required for Cortex Modification and Thus Spore Germination.mSphere. 2018 Jun 27;3(3):e00205-18. doi: 10.1128/mSphere.00205-18. Print 2018 Jun 27. mSphere. 2018. PMID: 29950380 Free PMC article.
-
Muramic lactam in peptidoglycan of Bacillus subtilis spores is required for spore outgrowth but not for spore dehydration or heat resistance.Proc Natl Acad Sci U S A. 1996 Dec 24;93(26):15405-10. doi: 10.1073/pnas.93.26.15405. Proc Natl Acad Sci U S A. 1996. PMID: 8986824 Free PMC article.
-
Clostridium difficile spore biology: sporulation, germination, and spore structural proteins.Trends Microbiol. 2014 Jul;22(7):406-16. doi: 10.1016/j.tim.2014.04.003. Epub 2014 May 7. Trends Microbiol. 2014. PMID: 24814671 Free PMC article. Review.
-
The role of peptidoglycan structure and structural dynamics during endospore dormancy and germination.Antonie Van Leeuwenhoek. 1999 May;75(4):299-307. doi: 10.1023/a:1001800507443. Antonie Van Leeuwenhoek. 1999. PMID: 10510717 Review.
Cited by
-
A cortex-specific penicillin-binding protein contributes to heat resistance in Clostridioides difficile spores.Anaerobe. 2021 Aug;70:102379. doi: 10.1016/j.anaerobe.2021.102379. Epub 2021 Apr 30. Anaerobe. 2021. PMID: 33940167 Free PMC article.
-
The small acid-soluble proteins of Clostridioides difficile regulate sporulation in a SpoIVB2-dependent manner.PLoS Pathog. 2024 Aug 30;20(8):e1012507. doi: 10.1371/journal.ppat.1012507. eCollection 2024 Aug. PLoS Pathog. 2024. PMID: 39213448 Free PMC article.
-
Reconstituting Spore Cortex Peptidoglycan Biosynthesis Reveals a Deacetylase That Catalyzes Transamidation.Biochemistry. 2023 Apr 18;62(8):1342-1346. doi: 10.1021/acs.biochem.3c00100. Epub 2023 Apr 6. Biochemistry. 2023. PMID: 37021938 Free PMC article.
-
A novel peptidoglycan deacetylase modulates daughter cell separation in E. coli.bioRxiv [Preprint]. 2025 Feb 19:2025.02.18.638797. doi: 10.1101/2025.02.18.638797. bioRxiv. 2025. PMID: 40027703 Free PMC article. Preprint.
-
Imaging Clostridioides difficile Spore Germination and Germination Proteins.J Bacteriol. 2022 Jul 19;204(7):e0021022. doi: 10.1128/jb.00210-22. Epub 2022 Jun 28. J Bacteriol. 2022. PMID: 35762766 Free PMC article.
References
-
- Cohen S. H., Gerding D. N., Johnson S., Kelly C. P., Loo V. G., McDonald L. C., Pepin J., Wilcox M. H., Society for Health care Epidemiology of America, and Infectious Diseases Society of America. (2010) Clinical practice guidelines for Clostridium difficile infection in adults: 2010 update by the society for health care epidemiology of America (SHEA) and the infectious diseases society of America (IDSA). Infect. Control Hosp. Epidemiol. 31, 431–455 10.1086/651706 - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

