Poly(ADP-ribose) polymerase 1 (PARP1) promotes oxidative stress-induced association of Cockayne syndrome group B protein with chromatin
- PMID: 30266807
- PMCID: PMC6240881
- DOI: 10.1074/jbc.RA118.004548
Poly(ADP-ribose) polymerase 1 (PARP1) promotes oxidative stress-induced association of Cockayne syndrome group B protein with chromatin
Abstract
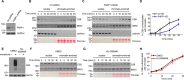
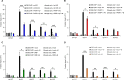
Cockayne syndrome protein B (CSB) is an ATP-dependent chromatin remodeler that relieves oxidative stress by regulating DNA repair and transcription. CSB is proposed to participate in base-excision repair (BER), the primary pathway for repairing oxidative DNA damage, but exactly how CSB participates in this process is unknown. It is also unclear whether CSB contributes to other repair pathways during oxidative stress. Here, using a patient-derived CS1AN-sv cell line, we examined how CSB is targeted to chromatin in response to menadione-induced oxidative stress, both globally and locus-specifically. We found that menadione-induced, global CSB-chromatin association does not require CSB's ATPase activity and is, therefore, mechanistically distinct from UV-induced CSB-chromatin association. Importantly, poly(ADP-ribose) polymerase 1 (PARP1) enhanced the kinetics of global menadione-induced CSB-chromatin association. We found that the major BER enzymes, 8-oxoguanine DNA glycosylase (OGG1) and apurinic/apyrimidinic endodeoxyribonuclease 1 (APE1), do not influence this association. Additionally, the level of γ-H2A histone family member X (γ-H2AX), a marker for dsDNA breaks, was not increased in menadione-treated cells. Therefore, our results support a model whereby PARP1 localizes to ssDNA breaks and recruits CSB to participate in DNA repair. Furthermore, this global CSB-chromatin association occurred independently of RNA polymerase II-mediated transcription elongation. However, unlike global CSB-chromatin association, both PARP1 knockdown and inhibition of transcription elongation interfered with menadione-induced CSB recruitment to specific genomic regions. This observation supports the hypothesis that CSB is also targeted to specific genomic loci to participate in transcriptional regulation in response to oxidative stress.
Keywords: ATP-dependent chromatin remodeling; CSB; Cockayne syndrome protein; DNA repair; PARP1; base-excision repair (BER); chromatin; oxidative stress; reactive oxygen species (ROS); single-strand DNA repair; transcription regulation.
© 2018 Boetefuer et al.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article
Figures


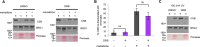


References
-
- Citterio E., Van Den Boom V., Schnitzler G., Kanaar R., Bonte E., Kingston R. E., Hoeijmakers J. H., and Vermeulen W. (2000) ATP-dependent chromatin remodeling by the Cockayne syndrome B DNA repair-transcription-coupling factor. Mol. Cell. Biol. 20, 7643–7653 10.1128/MCB.20.20.7643-7653.2000 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

