ASXL1 impairs osteoclast formation by epigenetic regulation of NFATc1
- PMID: 30266822
- PMCID: PMC6177649
- DOI: 10.1182/bloodadvances.2018018309
ASXL1 impairs osteoclast formation by epigenetic regulation of NFATc1
Abstract
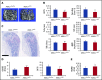
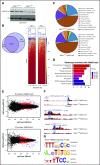
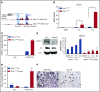
Additional sex comb-like 1 (ASXL1) mutations are commonly associated with myeloid malignancies and are markers of aggressive disease. The fact that ASXL1 is necessary for myeloid differentiation raises the possibility it also regulates osteoclasts. We find deletion of ASXL1 in myeloid cells results in bone loss with increased abundance of osteoclasts. Because ASXL1 is an enhancer of trithorax and polycomb (ETP) protein, we asked if it modulates osteoclast differentiation by maintaining balance between positive and negative epigenetic regulators. In fact, loss of ASXL1 induces concordant loss of inhibitory H3K27me3 with gain of H3K4me3 at key osteoclast differentiation genes, including nuclear factor for activated T cells 1 (NFATc1) and itgb3 In the setting of ASXL1 deficiency, increased NFATc1 binds to the Blimp1 (Prdm1) promoter thereby enhancing expression of this pro-osteoclastogenic gene. The global reduction of K27 trimethylation in ASXL1-deficient osteoclasts is also attended by a 40-fold increase in expression of the histone demethylase Jumonji domain-containing 3 (Jmjd3). Jmjd3 knockdown in ASXL1-deficient osteoclast precursors increases H3K27me3 on the NFATc1 promoter and impairs osteoclast formation. Thus, in addition to promoting myeloid malignancies, ASXL1 controls epigenetic reprogramming of osteoclasts to regulate bone resorption and mass.
© 2018 by The American Society of Hematology.
Conflict of interest statement
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Figures




References
-
- Novack DV, Teitelbaum SL. The osteoclast: friend or foe? Annu Rev Pathol. 2008;3(1):457-484. - PubMed
-
- Nishikawa K, Iwamoto Y, Kobayashi Y, et al. . DNA methyltransferase 3a regulates osteoclast differentiation by coupling to an S-adenosylmethionine-producing metabolic pathway. Nat Med. 2015;21(3):281-287. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

