Insights into a dual function amide oxidase/macrocyclase from lankacidin biosynthesis
- PMID: 30266997
- PMCID: PMC6162330
- DOI: 10.1038/s41467-018-06323-w
Insights into a dual function amide oxidase/macrocyclase from lankacidin biosynthesis
Erratum in
-
Author Correction: Insights into a dual function amide oxidase/macrocyclase from lankacidin biosynthesis.Nat Commun. 2019 Jan 29;10(1):553. doi: 10.1038/s41467-019-08571-w. Nat Commun. 2019. PMID: 30696828 Free PMC article.
-
Author Correction: Insights into a dual function amide oxidase/macrocyclase from lankacidin biosynthesis.Nat Commun. 2020 Jan 29;11(1):683. doi: 10.1038/s41467-020-14473-z. Nat Commun. 2020. PMID: 31996686 Free PMC article.
Abstract
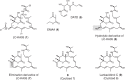
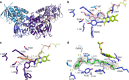
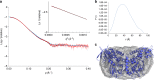
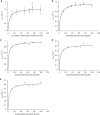
Acquisition of new catalytic activity is a relatively rare evolutionary event. A striking example appears in the pathway to the antibiotic lankacidin, as a monoamine oxidase (MAO) family member, LkcE, catalyzes both an unusual amide oxidation, and a subsequent intramolecular Mannich reaction to form the polyketide macrocycle. We report evidence here for the molecular basis for this dual activity. The reaction sequence involves several essential active site residues and a conformational change likely comprising an interdomain hinge movement. These features, which have not previously been described in the MAO family, both depend on a unique dimerization mode relative to all structurally characterized members. Taken together, these data add weight to the idea that designing new multifunctional enzymes may require changes in both architecture and catalytic machinery. Encouragingly, however, our data also show LkcE to bind alternative substrates, supporting its potential utility as a general cyclization catalyst in synthetic biology.
Conflict of interest statement
The authors declare no competing interests.
Figures


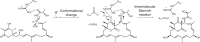
References
-
- Weiss, A. K. H., Loeffler, J. R., Liedl, K. R., Gstach, H. & Jansen-Dürr, P. The fumarylacetoacetate hydrolase (FAH) superfamily of enzymes: multifunctional enzymes from microbes to mitochondria. Biochem. Soc. Trans. 46, 295–309 (2018). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

