Simulations Reveal Multiple Intermediates in the Unzipping Mechanism of Neuronal SNARE Complex
- PMID: 30268539
- PMCID: PMC6260205
- DOI: 10.1016/j.bpj.2018.08.043
Simulations Reveal Multiple Intermediates in the Unzipping Mechanism of Neuronal SNARE Complex
Abstract
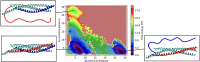
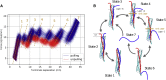
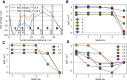
The assembling of the soluble N-ethylmaleimide-sensitive factor attachment protein receptor protein complex is a fundamental step in neuronal exocytosis, and it has been extensively studied in the last two decades. Yet, many details of this process remain inaccessible with the current experimental space and time resolution. Here, we study the zipping mechanism of the soluble N-ethylmaleimide-sensitive factor attachment protein receptor complex computationally by using a coarse-grained model. We explore the different pathways available and analyze their dependence on the computational model employed. We reveal and characterize multiple intermediate states, in agreement with previous experimental findings. We use our model to analyze the influence of single-residue mutations on the thermodynamics of the folding process.
Copyright © 2018 Biophysical Society. Published by Elsevier Inc. All rights reserved.
Figures


Similar articles
-
The influence of cell membrane and SNAP25 linker loop on the dynamics and unzipping of SNARE complex.PLoS One. 2017 Apr 20;12(4):e0176235. doi: 10.1371/journal.pone.0176235. eCollection 2017. PLoS One. 2017. PMID: 28426820 Free PMC article.
-
All-atom and coarse-grained simulations of the forced unfolding pathways of the SNARE complex.Proteins. 2014 Jul;82(7):1376-86. doi: 10.1002/prot.24505. Epub 2014 Feb 6. Proteins. 2014. PMID: 24403006
-
Structural transitions in the synaptic SNARE complex during Ca2+-triggered exocytosis.J Cell Biol. 2006 Jan 16;172(2):281-93. doi: 10.1083/jcb.200510012. J Cell Biol. 2006. PMID: 16418536 Free PMC article.
-
EPR Lineshape Analysis to Investigate the SNARE Folding Intermediates.Methods Mol Biol. 2019;1860:33-51. doi: 10.1007/978-1-4939-8760-3_3. Methods Mol Biol. 2019. PMID: 30317497 Free PMC article. Review.
-
Regulation of SNARE-mediated membrane fusion during exocytosis.Chem Rev. 2008 May;108(5):1669-86. doi: 10.1021/cr0782325. Epub 2008 Apr 18. Chem Rev. 2008. PMID: 18419164 Review. No abstract available.
References
-
- Söllner T., Whiteheart S.W., Rothman J.E. SNAP receptors implicated in vesicle targeting and fusion. Nature. 1993;362:318–324. - PubMed
-
- Weber T., Zemelman B.V., Rothman J.E. SNAREpins: minimal machinery for membrane fusion. Cell. 1998;92:759–772. - PubMed
-
- Sutton R.B., Fasshauer D., Brunger A.T. Crystal structure of a SNARE complex involved in synaptic exocytosis at 2.4 A resolution. Nature. 1998;395:347–353. - PubMed
-
- Südhof T.C. The synaptic vesicle cycle: a cascade of protein-protein interactions. Nature. 1995;375:645–653. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

