Deletion of the Mitochondrial Complex-IV Cofactor Heme A:Farnesyltransferase Causes Focal Segmental Glomerulosclerosis and Interferon Response
- PMID: 30268775
- PMCID: PMC6334261
- DOI: 10.1016/j.ajpath.2018.08.018
Deletion of the Mitochondrial Complex-IV Cofactor Heme A:Farnesyltransferase Causes Focal Segmental Glomerulosclerosis and Interferon Response
Abstract
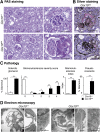
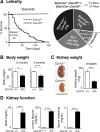
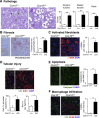
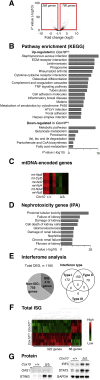
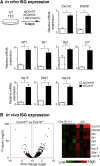
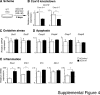
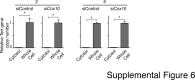
Mutations in mitochondrial DNA as well as nuclear-encoded mitochondrial proteins have been reported to cause tubulointerstitial kidney diseases and focal segmental glomerulosclerosis (FSGS). Recently, genes and pathways affecting mitochondrial turnover and permeability have been implicated in adult-onset FSGS. Furthermore, dysfunctioning mitochondria may be capable of engaging intracellular innate immune-sensing pathways. To determine the impact of mitochondrial dysfunction in FSGS and secondary innate immune responses, we generated Cre/loxP transgenic mice to generate a loss-of-function deletion mutation of the complex IV assembly cofactor heme A:farnesyltransferase (COX10) restricted to cells of the developing nephrons. These mice develop severe, early-onset FSGS with innate immune activation and die prematurely with kidney failure. Mutant kidneys showed loss of glomerular and tubular epithelial function, epithelial apoptosis, and, in addition, a marked interferon response. In vitro modeling of Cox10 deletion in primary kidney epithelium compromises oxygen consumption, ATP generation, and induces oxidative stress. In addition, loss of Cox10 triggers a selective interferon response, which may be caused by the leak of mitochondrial DNA into the cytosol activating the intracellular DNA sensor, stimulator of interferon genes. This new animal model provides a mechanism to study mitochondrial dysfunction in vivo and demonstrates a direct link between mitochondrial dysfunction and intracellular innate immune response.
Copyright © 2018 American Society for Investigative Pathology. Published by Elsevier Inc. All rights reserved.
Figures





Comment in
-
A link between mitochondrial dysfunction and innate immune activation in FSGS.Nat Rev Nephrol. 2018 Dec;14(12):721. doi: 10.1038/s41581-018-0075-6. Nat Rev Nephrol. 2018. PMID: 30327552 No abstract available.
References
-
- Dinour D., Mini S., Polak-Charcon S., Lotan D., Holtzman E.J. Progressive nephropathy associated with mitochondrial tRNA gene mutation. Clin Nephrol. 2004;62:149–154. - PubMed
-
- Finsterer J. Mitochondriopathies. Eur J Neurol. 2004;11:163–186. - PubMed
-
- Niaudet P., Rotig A. The kidney in mitochondrial cytopathies. Kidney Int. 1997;51:1000–1007. - PubMed
-
- Genovese G., Friedman D.J., Ross M.D., Lecordier L., Uzureau P., Freedman B.I., Bowden D.W., Langefeld C.D., Oleksyk T.K., Uscinski Knob A.L., Bernhardy A.J., Hicks P.J., Nelson G.W., Vanhollebeke B., Winkler C.A., Kopp J.B., Pays E., Pollak M.R. Association of trypanolytic ApoL1 variants with kidney disease in African Americans. Science. 2010;329:841–845. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

