Probe-free label system for rapid detection of Cronobacter genus in powdered infant formula
- PMID: 30269246
- PMCID: PMC6163125
- DOI: 10.1186/s13568-018-0689-x
Probe-free label system for rapid detection of Cronobacter genus in powdered infant formula
Abstract
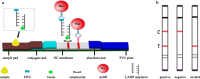
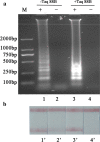
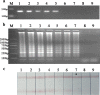
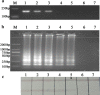
Cronobacter species previously known as Enterobacter sakazakii poses high risks to neonates and infants. In this work a rapid detection method was developed which combined loop-mediated isothermal amplification with lateral flow assay for detection of Cronobacter species in powdered infant formula. The fast amplification reaction without betaine was established and capable of performing DNA replication within 25 min. Based on the novel probe-free labeling methods, we established a lateral flow assay to capture the specific loop-mediated isothermal amplification amplicons which were labeled with fluorescein isothiocyanate and biotin. And the final detection time of this system was within 40 min. The false positive results of the lateral flow assay induced by primer dimer tagged with fluorescein isothiocyanate and biotin were eliminated by Taq single strand DNA binding protein (4 ng/μL). Simultaneously, the efficiency of the fast loop-mediated isothermal amplification assay was achieved. By injection of Taq SSB into the amplification assay as a replacement for betaine, the novel probe-free method could detect Cronobacter species with high specificity and sensitivity at the detection limit in PIF of 101 cfu/g. Our overall strategy has excellent potential in the rapid diagnosis of Cronobacter species label-free by integrating loop-mediated isothermal amplification and lateral flow assay.
Keywords: Cronobacter species; Loop-mediated isothermal amplification; Powdered infant formula; Probe-free label based lateral flow assay.
Figures


Similar articles
-
Screening of specific nucleic acid targets for Cronobacter sakazakii and visual detection by loop-mediated isothermal amplification and lateral flow dipstick method in powdered infant formula.J Dairy Sci. 2021 May;104(5):5152-5165. doi: 10.3168/jds.2020-19427. Epub 2021 Mar 2. J Dairy Sci. 2021. PMID: 33663822
-
Multiplex loop-mediated isothermal amplification-based lateral flow dipstick for simultaneous detection of 3 food-borne pathogens in powdered infant formula.J Dairy Sci. 2020 May;103(5):4002-4012. doi: 10.3168/jds.2019-17538. Epub 2020 Feb 26. J Dairy Sci. 2020. PMID: 32113770
-
Electricity-free amplification and visual detection of Cronobacter species in powdered infant formula.J Dairy Sci. 2020 Aug;103(8):6882-6893. doi: 10.3168/jds.2019-17661. Epub 2020 Jun 3. J Dairy Sci. 2020. PMID: 32505404
-
Cronobacter species (formerly known as Enterobacter sakazakii) in powdered infant formula: a review of our current understanding of the biology of this bacterium.J Appl Microbiol. 2012 Jul;113(1):1-15. doi: 10.1111/j.1365-2672.2012.05281.x. Epub 2012 Apr 11. J Appl Microbiol. 2012. PMID: 22420458 Review.
-
Cronobacter Species Contamination of Powdered Infant Formula and the Implications for Neonatal Health.Front Pediatr. 2015 Jul 2;3:56. doi: 10.3389/fped.2015.00056. eCollection 2015. Front Pediatr. 2015. PMID: 26191519 Free PMC article. Review.
Cited by
-
Establishment of Sample-to-Answer Loop-Mediated Isothermal Amplification-Based Nucleic Acid Testing Using the Sampling, Processing, Incubation, Detection and Lateral Flow Immunoassay Platforms.Biosensors (Basel). 2024 Dec 13;14(12):609. doi: 10.3390/bios14120609. Biosensors (Basel). 2024. PMID: 39727874 Free PMC article.
-
Immuno-Dipstick for Colletotrichum gloeosporioides Detection: Towards On-Farm Application.Biosensors (Basel). 2022 Jan 18;12(2):49. doi: 10.3390/bios12020049. Biosensors (Basel). 2022. PMID: 35200310 Free PMC article.
-
Vigilant: An Engineered VirD2-Cas9 Complex for Lateral Flow Assay-Based Detection of SARS-CoV2.Nano Lett. 2021 Apr 28;21(8):3596-3603. doi: 10.1021/acs.nanolett.1c00612. Epub 2021 Apr 12. Nano Lett. 2021. PMID: 33844549 Free PMC article.
-
Lateral Flow Recombinase Polymerase Amplification Assays for the Detection of Human Plasmodium Species.Am J Trop Med Hyg. 2023 Mar 13;108(5):882-886. doi: 10.4269/ajtmh.22-0657. Print 2023 May 3. Am J Trop Med Hyg. 2023. PMID: 36913921 Free PMC article.
-
Clinical testing on SARS-CoV-2 swab samples using reverse-transcription loop-mediated isothermal amplification (RT-LAMP).BMC Infect Dis. 2022 Aug 18;22(1):697. doi: 10.1186/s12879-022-07684-w. BMC Infect Dis. 2022. PMID: 35982419 Free PMC article.
References
-
- Blažková M, Koets M, Rauch P, Amerongen AV. Development of a nucleic acid lateral flow immunoassay for simultaneous detection of Listeria spp. and Listeria monocytogenes in food. Eur Food Res Technol. 2009;229(6):867–874. doi: 10.1007/s00217-009-1115-z. - DOI
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

