ARL3 Mutations Cause Joubert Syndrome by Disrupting Ciliary Protein Composition
- PMID: 30269812
- PMCID: PMC6174286
- DOI: 10.1016/j.ajhg.2018.08.015
ARL3 Mutations Cause Joubert Syndrome by Disrupting Ciliary Protein Composition
Abstract
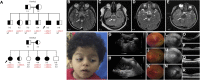
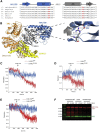
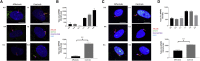
Joubert syndrome (JBTS) is a genetically heterogeneous autosomal-recessive neurodevelopmental ciliopathy. We investigated further the underlying genetic etiology of Joubert syndrome by studying two unrelated families in whom JBTS was not associated with pathogenic variants in known JBTS-associated genes. Combined autozygosity mapping of both families highlighted a candidate locus on chromosome 10 (chr10: 101569997-109106128, UCSC Genome Browser hg 19), and exome sequencing revealed two missense variants in ARL3 within the candidate locus. The encoded protein, ADP ribosylation factor-like GTPase 3 (ARL3), is a small GTP-binding protein that is involved in directing lipid-modified proteins into the cilium in a GTP-dependent manner. Both missense variants replace the highly conserved Arg149 residue, which we show to be necessary for the interaction with its guanine nucleotide exchange factor ARL13B, such that the mutant protein is associated with reduced INPP5E and NPHP3 localization in cilia. We propose that ARL3 provides a potential hub in the network of proteins implicated in ciliopathies, whereby perturbation of ARL3 leads to the mislocalization of multiple ciliary proteins as a result of abnormal displacement of lipidated protein cargo.
Keywords: ARL13B; ARL3; Joubert syndrome; cilia; guanine nucleotide exchange factor; trafficking.
Copyright © 2018 The Author(s). Published by Elsevier Inc. All rights reserved.
Figures


References
-
- Srivastava S., Ramsbottom S.A., Molinari E., Alkanderi S., Filby A., White K., Henry C., Saunier S., Miles C.G., Sayer J.A. A human patient-derived cellular model of Joubert syndrome reveals ciliary defects which can be rescued with targeted therapies. Hum. Mol. Genet. 2017;26:4657–4667. - PMC - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

