RAB11FIP5 Expression and Altered Natural Killer Cell Function Are Associated with Induction of HIV Broadly Neutralizing Antibody Responses
- PMID: 30270043
- PMCID: PMC6176872
- DOI: 10.1016/j.cell.2018.08.064
RAB11FIP5 Expression and Altered Natural Killer Cell Function Are Associated with Induction of HIV Broadly Neutralizing Antibody Responses
Abstract
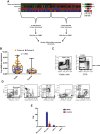
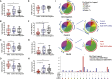
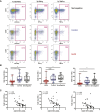
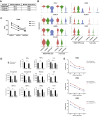
HIV-1 broadly neutralizing antibodies (bnAbs) are difficult to induce with vaccines but are generated in ∼50% of HIV-1-infected individuals. Understanding the molecular mechanisms of host control of bnAb induction is critical to vaccine design. Here, we performed a transcriptome analysis of blood mononuclear cells from 47 HIV-1-infected individuals who made bnAbs and 46 HIV-1-infected individuals who did not and identified in bnAb individuals upregulation of RAB11FIP5, encoding a Rab effector protein associated with recycling endosomes. Natural killer (NK) cells had the highest differential expression of RAB11FIP5, which was associated with greater dysregulation of NK cell subsets in bnAb subjects. NK cells from bnAb individuals had a more adaptive/dysfunctional phenotype and exhibited impaired degranulation and cytokine production that correlated with RAB11FIP5 transcript levels. Moreover, RAB11FIP5 overexpression modulated the function of NK cells. These data suggest that NK cells and Rab11 recycling endosomal transport are involved in regulation of HIV-1 bnAb development.
Keywords: HIV-1; Rab11fip5; broadly neutralizing antibodies; natural killer cells; recycling endosomes; vaccine.
Copyright © 2018 The Authors. Published by Elsevier Inc. All rights reserved.
Figures










References
-
- Alter G., Teigen N., Davis B.T., Addo M.M., Suscovich T.J., Waring M.T., Streeck H., Johnston M.N., Staller K.D., Zaman M.T. Sequential deregulation of NK cell subset distribution and function starting in acute HIV-1 infection. Blood. 2005;106:3366–3369. - PubMed
-
- Björkström N.K., Ljunggren H.G., Sandberg J.K. CD56 negative NK cells: origin, function, and role in chronic viral disease. Trends Immunol. 2010;31:401–406. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

