Age-dependent methylation in epigenetic clock CpGs is associated with G-quadruplex, co-transcriptionally formed RNA structures and tentative splice sites
- PMID: 30270726
- PMCID: PMC6224212
- DOI: 10.1080/15592294.2018.1514232
Age-dependent methylation in epigenetic clock CpGs is associated with G-quadruplex, co-transcriptionally formed RNA structures and tentative splice sites
Abstract
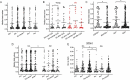
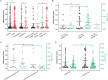
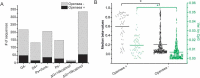
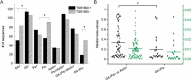
Horvath's epigenetic clock consists of 353 CpGs whose methylation levels can accurately predict the age of individuals. Using bioinformatics analysis, we investigated the conformation, energy characteristics and presence of tentative splice sites of the sequences surrounding the epigenetic clock CpGs, in relation to the median methylation changes in different ages, the presence of CpG islands and their position in genes. Common characteristics in the 100 nt sequences surrounding the epigenetic clock CpGs are G-quadruplexes and/or tentative splice site motifs. Median methylation increases significantly in sequences which adopt less stable structures during transcription. Methylation is higher when CpGs overlap with G-quadruplexes than when they precede them. Median methylation in epigenetic clock CpGs is higher in sequences expressed as single products rather than in multiple products and those containing single donors and multiple acceptors. Age-related methylation variation is significant in sequences without G-quadruplexes, particularly those producing low stability nascent RNA and those with splice sites. CpGs in sequences close to transcription start sites and those which are possibly never expressed (hypothetical proteins) undergo similar extent of age-related median methylation decrease and increase. Preservation of methylation is observed in CpG islands without G-quadruplexes, contrary to CpGs far from CpG islands (open sea). Sequences containing G-quadruplexes and RNA pseudoknots, determining the recognition by H3K27 histone methyltransferase, are hypomethylated. The presented structural DNA and co-transcriptional RNA analysis of epigenetic clock sequences, foreshadows the association of age-related methylation changes with the principle biological processes of DNA and histone methylation, splicing and chromatin silencing.
Keywords: DNA methylation; G-quadruplex; aging; alternative splicing; co-transcriptionally formed RNA structures; epigenetic clock; hypothetical proteins; tentative splice sites.
Figures


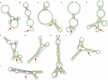
References
-
- Rönn T, Volkov P, Gillberg L, et al. Impact of age, BMI and HbA1c levels on the genome-wide DNA methylation and mRNA expression patterns in human adipose tissue and identification of epigenetic biomarkers in blood. Hum Mol Genet. 2015;24:3792–3813. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
