Single Nucleotide Polymorphism Analysis Indicates Genetic Distinction and Reduced Diversity of Swine-Associated Methicillin Resistant Staphylococcus aureus (MRSA) ST5 Isolates Compared to Clinical MRSA ST5 Isolates
- PMID: 30271385
- PMCID: PMC6142820
- DOI: 10.3389/fmicb.2018.02078
Single Nucleotide Polymorphism Analysis Indicates Genetic Distinction and Reduced Diversity of Swine-Associated Methicillin Resistant Staphylococcus aureus (MRSA) ST5 Isolates Compared to Clinical MRSA ST5 Isolates
Abstract
Livestock associated methicillin resistant S. aureus (LA-MRSA) are lineages adapted to livestock species. LA-MRSA can be transmitted to humans and public health concerns exist because livestock may be the largest MRSA reservoir outside of hospital settings. Although the predominant European (ST398) and Asian (ST9) lineages of LA-MRSA are considered livestock adapted, North American swine also harbor ST5, a globally disseminated and highly pathogenic lineage. This study applied whole genome sequencing and single nucleotide polymorphism (SNP) typing to compare the population structure and genetic relatedness between swine associated and human clinical MRSA ST5 isolates. The established high-resolution phylogenomic framework revealed that LA-MRSA and human clinical MRSA ST5 are genetically distinct. LA-MRSA isolates were found to be clonal within farms, while greater genome diversity was observed among sampled clinical MRSA ST5. Analysis of the accessory genome demonstrated that LA-MRSA ST5 isolates and clinical MRSA ST5 isolates harbor different AMR genes and virulence factors, consistent with the SNP analysis. Collectively, our data indicate LA-MRSA and clinical MRSA ST5 isolates are distinct and the swine reservoir is likely of minimal significance as a source of clinical MRSA ST5 infections.
Keywords: LA-MRSA; Staphylococcus aureus; agriculture; mobile genetic elements; phylogenetic analysis; single nucleotide polymorphism (SNP) typing; swine; whole genome sequence (WGS).
Figures

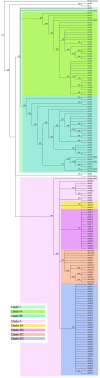
References
-
- Andreoletti O., Budka H., Buncic S., Colin P., Collins J. D., De Koeijer A., et al. (2009). Scientific opinion of the panel on biological hazards on a request from the European commission on assessment of the public health significance of meticillin resistant Staphylococcus aureus (MRSA) in animals and foods. EFSA J. 993 1–73.
-
- Bal A. M., Coombs G. W., Holden M. T., Lindsay J. A., Nimmo G. R., Tattevin P., et al. (2016). Genomic insights into the emergence and spread of international clones of healthcare-, community- and livestock-associated meticillin-resistant Staphylococcus aureus: blurring of the traditional definitions. J Glob Antimicrob Resist 6 95–101. 10.1016/j.jgar.2016.04.004 - DOI - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

