Prediction Is a Balancing Act: Importance of Sampling Methods to Balance Sensitivity and Specificity of Predictive Models Based on Imbalanced Chemical Data Sets
- PMID: 30271769
- PMCID: PMC6149243
- DOI: 10.3389/fchem.2018.00362
Prediction Is a Balancing Act: Importance of Sampling Methods to Balance Sensitivity and Specificity of Predictive Models Based on Imbalanced Chemical Data Sets
Abstract
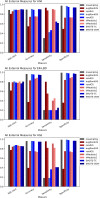
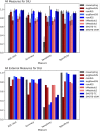
Increase in the number of new chemicals synthesized in past decades has resulted in constant growth in the development and application of computational models for prediction of activity as well as safety profiles of the chemicals. Most of the time, such computational models and its application must deal with imbalanced chemical data. It is indeed a challenge to construct a classifier using imbalanced data set. In this study, we analyzed and validated the importance of different sampling methods over non-sampling method, to achieve a well-balanced sensitivity and specificity of a machine learning model trained on imbalanced chemical data. Additionally, this study has achieved an accuracy of 93.00%, an AUC of 0.94, F1 measure of 0.90, sensitivity of 96.00% and specificity of 91.00% using SMOTE sampling and Random Forest classifier for the prediction of Drug Induced Liver Injury (DILI). Our results suggest that, irrespective of data set used, sampling methods can have major influence on reducing the gap between sensitivity and specificity of a model. This study demonstrates the efficacy of different sampling methods for class imbalanced problem using binary chemical data sets.
Keywords: DILI; SMOTE; Tox21; imbalanced data; machine learning; molecular fingerprints; sampling methods; sensitivity-specificity balance.
Figures



References
-
- Beyan C., Fisher R. (2015). Classifying imbalanced data sets using similarity based hierarchical decomposition. Pattern Recognit. 48, 1653–1672. 10.1016/j.patcog.2014.10.032 - DOI
-
- Capuzzi S. J., Regina P., Isayev O., Farag S., Tropsha A. (2016). QSAR modeling of Tox21 challenge stress response and nuclear receptor signaling toxicity assays. Front. Environ. Sci. 4:3 10.3389/fenvs.2016.00003 - DOI
LinkOut - more resources
Full Text Sources
Miscellaneous

