Acidity promotes degradation of multi-species environmental DNA in lotic mesocosms
- PMID: 30271891
- PMCID: PMC6123786
- DOI: 10.1038/s42003-017-0005-3
Acidity promotes degradation of multi-species environmental DNA in lotic mesocosms
Abstract
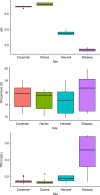
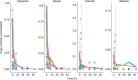
Accurate quantification of biodiversity is fundamental to understanding ecosystem function and for environmental assessment. Molecular methods using environmental DNA (eDNA) offer a non-invasive, rapid, and cost-effective alternative to traditional biodiversity assessments, which require high levels of expertise. While eDNA analyses are increasingly being utilized, there remains considerable uncertainty regarding the dynamics of multispecies eDNA, especially in variable systems such as rivers. Here, we utilize four sets of upland stream mesocosms, across an acid-base gradient, to assess the temporal and environmental degradation of multispecies eDNA. Sampling included water column and biofilm sampling over time with eDNA quantified using qPCR. Our findings show that the persistence of lotic multispecies eDNA, sampled from water and biofilm, decays to non-detectable levels within 2 days and that acidic environments accelerate the degradation process. Collectively, the results provide the basis for a predictive framework for the relationship between lotic eDNA degradation dynamics in spatio-temporally dynamic river ecosystems.
Conflict of interest statement
The authors declare no competing financial interests.
Figures


References
-
- Barnes MA, Turner CR. The ecology of environmental DNA and implications for conservation genetics. Conserv. Genet. 2016;17:1–17. doi: 10.1007/s10592-015-0775-4. - DOI
-
- Jerde CL, Mahon AR, Chadderton WL, Lodge DM. ‘Sight-unseen’ detection of rare aquatic species using environmental DNA. Conserv. Lett. 2011;4:150–157. doi: 10.1111/j.1755-263X.2010.00158.x. - DOI
-
- Biggs J, et al. Using eDNA to develop a national citizen science-based monitoring programme for the great crested newt (Triturus cristatus) Biol. Conserv. 2015;183:19–28. doi: 10.1016/j.biocon.2014.11.029. - DOI
-
- Thomsen PF, Willerslev E. Environmental DNA – an emerging tool in conservation for monitoring past and present biodiversity. Biol. Conserv. 2015;183:4–18. doi: 10.1016/j.biocon.2014.11.019. - DOI

