The GTPase domain of gamma-tubulin is required for normal mitochondrial function and spatial organization
- PMID: 30271923
- PMCID: PMC6123723
- DOI: 10.1038/s42003-018-0037-3
The GTPase domain of gamma-tubulin is required for normal mitochondrial function and spatial organization
Abstract
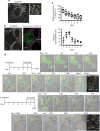
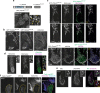
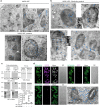
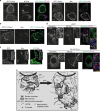
In the cell, γ-tubulin establishes a cellular network of threads named the γ-string meshwork. However, the functions of this meshwork remain to be determined. We investigated the traits of the meshwork and show that γ-strings have the ability to connect the cytoplasm and the mitochondrial DNA together. We also show that γ-tubulin has a role in the maintenance of the mitochondrial network and functions as reduced levels of γ-tubulin or impairment of its GTPase domain disrupts the mitochondrial network and alters both their respiratory capacity and the expression of mitochondrial-related genes. By contrast, reduced mitochondrial number or increased protein levels of γ-tubulin DNA-binding domain enhanced the association of γ-tubulin with mitochondria. Our results demonstrate that γ-tubulin is an important mitochondrial structural component that maintains the mitochondrial network, providing mitochondria with a cellular infrastructure. We propose that γ-tubulin provides a cytoskeletal element that gives form to the mitochondrial network.
Conflict of interest statement
The authors declare no competing interests.
Figures





References
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

