Sea turtle fibropapilloma tumors share genomic drivers and therapeutic vulnerabilities with human cancers
- PMID: 30271945
- PMCID: PMC6123702
- DOI: 10.1038/s42003-018-0059-x
Sea turtle fibropapilloma tumors share genomic drivers and therapeutic vulnerabilities with human cancers
Abstract
Wildlife populations are under intense anthropogenic pressures, with the geographic range of many species shrinking, dramatic reductions in population numbers and undisturbed habitats, and biodiversity loss. It is postulated that we are in the midst of a sixth (Anthropocene) mass extinction event, the first to be induced by human activity. Further, threatening vulnerable species is the increased rate of emerging diseases, another consequence of anthropogenic activities. Innovative approaches are required to help maintain healthy populations until the chronic underlying causes of these issues can be addressed. Fibropapillomatosis in sea turtles is one such wildlife disease. Here, we applied precision-medicine-based approaches to profile fibropapillomatosis tumors to better understand their biology, identify novel therapeutics, and gain insights into viral and environmental triggers for fibropapillomatosis. We show that fibropapillomatosis tumors share genetic vulnerabilities with human cancer types, revealing that they are amenable to treatment with human anti-cancer therapeutics.
Conflict of interest statement
The authors declare no competing interests.
Figures


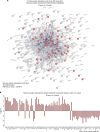
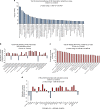



References
-
- Smith GC, Coates CW. Fibro-epithelial growths of the skin in large marine turtles Chelonia mydas. Zoologica. 1938;23:93–98.
-
- Lucke B. Studies on tumors in cold-blooded vertebrates. Annual Report of the Tortugas Laboratory of the Camegie Institute. 1938;1937:92–94.
-
- Cruz Sr, E. (1985). Saga of the sea turtle. FL, USA: Privately-published.
LinkOut - more resources
Full Text Sources

