Unusual dicistronic expression from closely spaced initiation codons in an umbravirus subgenomic RNA
- PMID: 30272199
- PMCID: PMC6294492
- DOI: 10.1093/nar/gky871
Unusual dicistronic expression from closely spaced initiation codons in an umbravirus subgenomic RNA
Abstract
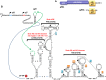
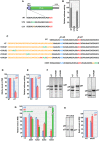
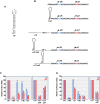
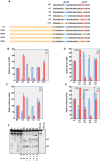
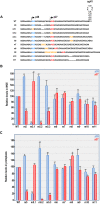
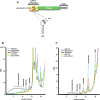
Translation commencing at closely spaced initiation codons is common in RNA viruses with limited genome space. In the subgenomic RNA (sgRNA) of Pea enation mosaic virus 2, two closely spaced, out-of-frame start codons direct synthesis of movement/stability proteins p26 and p27. Efficient translation from AUG26/AUG27 is dependent on three 3'-proximal cap-independent translation enhancers (3'CITEs), whereas translation of the genomic (gRNA) requires only two. Contrary to strictly scanning-dependent initiation at the gRNA, sequence context of AUG26/AUG27 does not conform with Kozak requirements and insertion of efficient upstream AUGs had pronounced effects for AUG26 but only moderate effects for AUG27. Insertion of a hairpin within an extended 5' UTR did not significantly impact translation from AUG26/AUG27. Furthermore, AUG27 repressed translation from upstream AUG26 and this effect was mitigated when inter-codon spacing was reduced. Addition of a stable hairpin to the very 5' end of the sgRNA severely restricted translation, testifying that this 3'CITE-driven initiation is 5' end-dependent. Similar to gRNA, sgRNA reporter transcripts were nearly exclusively associated with light polysomes and 3'CITE-promoted long-distance interaction connecting the sgRNA ends affected the number of templates translated and not the initiation rate. We propose a non-canonical, 3'CITE-driven mechanism for efficient dicistronic expression from umbravirus sgRNAs.
Figures


Similar articles
-
Conserved Structure Associated with Different 3'CITEs Is Important for Translation of Umbraviruses.Viruses. 2023 Feb 27;15(3):638. doi: 10.3390/v15030638. Viruses. 2023. PMID: 36992347 Free PMC article.
-
Identification of Novel 5' and 3' Translation Enhancers in Umbravirus-Like Coat Protein-Deficient RNA Replicons.J Virol. 2022 Apr 13;96(7):e0173621. doi: 10.1128/jvi.01736-21. Epub 2022 Mar 17. J Virol. 2022. PMID: 35297668 Free PMC article.
-
The 3' untranslated region of Pea Enation Mosaic Virus contains two T-shaped, ribosome-binding, cap-independent translation enhancers.J Virol. 2014 Oct;88(20):11696-712. doi: 10.1128/JVI.01433-14. Epub 2014 Aug 6. J Virol. 2014. PMID: 25100834 Free PMC article.
-
Cis- and trans-regulation of luteovirus gene expression by the 3' end of the viral genome.Virus Res. 2015 Aug 3;206:37-45. doi: 10.1016/j.virusres.2015.03.009. Epub 2015 Apr 6. Virus Res. 2015. PMID: 25858272 Free PMC article. Review.
-
3' Cap-independent translation enhancers of positive-strand RNA plant viruses.Curr Opin Virol. 2011 Nov;1(5):373-80. doi: 10.1016/j.coviro.2011.10.002. Epub 2011 Oct 24. Curr Opin Virol. 2011. PMID: 22440838 Review.
Cited by
-
The Multifunctional Long-Distance Movement Protein of Pea Enation Mosaic Virus 2 Protects Viral and Host Transcripts from Nonsense-Mediated Decay.mBio. 2020 Mar 10;11(2):e00204-20. doi: 10.1128/mBio.00204-20. mBio. 2020. PMID: 32156817 Free PMC article.
-
Opium Poppy Mosaic Virus Has an Xrn-Resistant, Translated Subgenomic RNA and a BTE 3' CITE.J Virol. 2021 Apr 12;95(9):e02109-20. doi: 10.1128/JVI.02109-20. Print 2021 Apr 12. J Virol. 2021. PMID: 33597210 Free PMC article.
-
Functional Cyclization of Eukaryotic mRNAs.Int J Mol Sci. 2020 Feb 29;21(5):1677. doi: 10.3390/ijms21051677. Int J Mol Sci. 2020. PMID: 32121426 Free PMC article.
-
Conserved Structure Associated with Different 3'CITEs Is Important for Translation of Umbraviruses.Viruses. 2023 Feb 27;15(3):638. doi: 10.3390/v15030638. Viruses. 2023. PMID: 36992347 Free PMC article.
-
Umbravirus-like RNA viruses are capable of independent systemic plant infection in the absence of encoded movement proteins.PLoS Biol. 2024 Apr 25;22(4):e3002600. doi: 10.1371/journal.pbio.3002600. eCollection 2024 Apr. PLoS Biol. 2024. PMID: 38662792 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

