The DEAD-box RNA-binding protein DDX6 regulates parental RNA decay for cellular reprogramming to pluripotency
- PMID: 30273347
- PMCID: PMC6166933
- DOI: 10.1371/journal.pone.0203708
The DEAD-box RNA-binding protein DDX6 regulates parental RNA decay for cellular reprogramming to pluripotency
Abstract
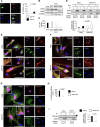
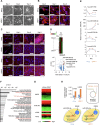
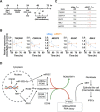
Cellular transitions and differentiation processes require mRNAs supporting the new phenotype but also the clearance of existing mRNAs for the parental phenotype. Cellular reprogramming from fibroblasts to induced pluripotent stem cells (iPSCs) occurs at the early stage of mesenchymal epithelial transition (MET) and involves drastic morphological changes. We examined the molecular mechanism for MET, focusing on RNA metabolism. DDX6, an RNA helicase, was indispensable for iPSC formation, in addition to RO60 and RNY1, a non-coding RNA, which form complexes involved in intracellular nucleotide sensing. RO60/RNY1/DDX6 complexes formed prior to processing body formation, which is central to RNA metabolism. The abrogation of DDX6 expression inhibited iPSC generation, which was mediated by RNA decay targeting parental mRNAs supporting mesenchymal phenotypes, along with microRNAs, such as miR-302b-3p. These results show that parental mRNA clearance is a prerequisite for cellular reprogramming and that DDX6 plays a central role in this process.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


References
-
- Clark G, Reichlin M, Tomasi TB Jr. Characterization of a soluble cytoplasmic antigen reactive with sera from patients with systemic lupus erythmatosus. J Immunol. 1969;102(1):117–22. . - PubMed
-
- Lerner MR, Boyle JA, Hardin JA, Steitz JA. Two novel classes of small ribonucleoproteins detected by antibodies associated with lupus erythematosus. Science. 1981;211(4480):400–2. Epub 1981/01/23. . - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

