Phylogenetically Novel Uncultured Microbial Cells Dominate Earth Microbiomes
- PMID: 30273414
- PMCID: PMC6156271
- DOI: 10.1128/mSystems.00055-18
Phylogenetically Novel Uncultured Microbial Cells Dominate Earth Microbiomes
Abstract
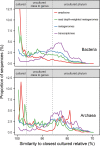
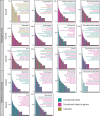
To describe a microbe's physiology, including its metabolism, environmental roles, and growth characteristics, it must be grown in a laboratory culture. Unfortunately, many phylogenetically novel groups have never been cultured, so their physiologies have only been inferred from genomics and environmental characteristics. Although the diversity, or number of different taxonomic groups, of uncultured clades has been studied well, their global abundances, or numbers of cells in any given environment, have not been assessed. We quantified the degree of similarity of 16S rRNA gene sequences from diverse environments in publicly available metagenome and metatranscriptome databases, which we show have far less of the culture bias present in primer-amplified 16S rRNA gene surveys, to those of their nearest cultured relatives. Whether normalized to scaffold read depths or not, the highest abundances of metagenomic 16S rRNA gene sequences belong to phylogenetically novel uncultured groups in seawater, freshwater, terrestrial subsurface, soil, hypersaline environments, marine sediment, hot springs, hydrothermal vents, nonhuman hosts, snow, and bioreactors (22% to 87% uncultured genera to classes and 0% to 64% uncultured phyla). The exceptions were human and human-associated environments, which were dominated by cultured genera (45% to 97%). We estimate that uncultured genera and phyla could comprise 7.3 × 1029 (81%) and 2.2 × 1029 (25%) of microbial cells, respectively. Uncultured phyla were overrepresented in metatranscriptomes relative to metagenomes (46% to 84% of sequences in a given environment), suggesting that they are viable. Therefore, uncultured microbes, often from deeply phylogenetically divergent groups, dominate nonhuman environments on Earth, and their undiscovered physiologies may matter for Earth systems. IMPORTANCE In the past few decades, it has become apparent that most of the microbial diversity on Earth has never been characterized in laboratory cultures. We show that these unknown microbes, sometimes called "microbial dark matter," are numerically dominant in all major environments on Earth, with the exception of the human body, where most of the microbes have been cultured. We also estimate that about one-quarter of the population of microbial cells on Earth belong to phyla with no cultured relatives, suggesting that these never-before-studied organisms may be important for ecosystem functions. Author Video: An author video summary of this article is available.
Keywords: environmental microbiology; phylogeny; uncultured microbes.
Figures


Comment in
-
The majority is uncultured.Nat Rev Microbiol. 2018 Dec;16(12):716-717. doi: 10.1038/s41579-018-0097-x. Nat Rev Microbiol. 2018. PMID: 30275521 No abstract available.
References
-
- Rinke C, Schwientek P, Sczyrba A, Ivanova NN, Anderson IJ, Cheng J-F, Darling A, Malfatti S, Swan BK, Gies EA, Dodsworth JA, Hedlund BP, Tsiamis G, Sievert SM, Liu W-T, Eisen JA, Hallam SJ, Kyrpides NC, Stepanauskas R, Rubin EM, Hugenholtz P, Woyke T. 2013. Insights into the phylogeny and coding potential of microbial dark matter. Nature 499:431–437. doi:10.1038/nature12352. - DOI - PubMed
-
- Wang Y, Hatt JK, Tsementzi D, Rodriguez-R LM, Ruiz-Perez CA, Weigand MR, Kizer H, Maresca G, Krishnan R, Poretsky R, Spain JC, Konstantinidis KT. 2017. Quantifying the importance of the rare biosphere for microbial community response to organic pollutants in a freshwater ecosystem. Appl Environ Microbiol 83:1–19. doi:10.1128/AEM.03321-16. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

