Epithelial RNase H2 Maintains Genome Integrity and Prevents Intestinal Tumorigenesis in Mice
- PMID: 30273559
- PMCID: PMC6311085
- DOI: 10.1053/j.gastro.2018.09.047
Epithelial RNase H2 Maintains Genome Integrity and Prevents Intestinal Tumorigenesis in Mice
Abstract
Background & aims: RNase H2 is a holoenzyme, composed of 3 subunits (ribonuclease H2 subunits A, B, and C), that cleaves RNA:DNA hybrids and removes mis-incorporated ribonucleotides from genomic DNA through ribonucleotide excision repair. Ribonucleotide incorporation by eukaryotic DNA polymerases occurs during every round of genome duplication and produces the most frequent type of naturally occurring DNA lesion. We investigated whether intestinal epithelial proliferation requires RNase H2 function and whether RNase H2 activity is disrupted during intestinal carcinogenesis.
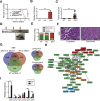
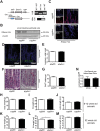
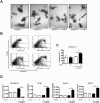
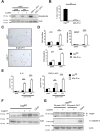
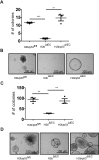
Methods: We generated mice with epithelial-specific deletion of ribonuclease H2 subunit B (H2bΔIEC) and mice that also had deletion of tumor-suppressor protein p53 (H2b/p53ΔIEC); we compared phenotypes with those of littermate H2bfl/fl or H2b/p53fl/fl (control) mice at young and old ages. Intestinal tissues were collected and analyzed by histology. We isolated epithelial cells, generated intestinal organoids, and performed RNA sequence analyses. Mutation signatures of spontaneous tumors from H2b/p53ΔIEC mice were characterized by exome sequencing. We collected colorectal tumor specimens from 467 patients, measured levels of ribonuclease H2 subunit B, and associated these with patient survival times and transcriptome data.
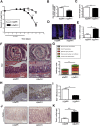
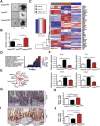
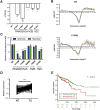
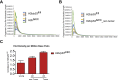
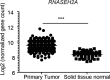
Results: The H2bΔIEC mice had DNA damage to intestinal epithelial cells and proliferative exhaustion of the intestinal stem cell compartment compared with controls and H2b/p53ΔIEC mice. However, H2b/p53ΔIEC mice spontaneously developed small intestine and colon carcinomas. DNA from these tumors contained T>G base substitutions at GTG trinucleotides. Analyses of transcriptomes of human colorectal tumors associated lower levels of RNase H2 with shorter survival times.
Conclusions: In analyses of mice with disruption of the ribonuclease H2 subunit B gene and colorectal tumors from patients, we provide evidence that RNase H2 functions as a colorectal tumor suppressor. H2b/p53ΔIEC mice can be used to study the roles of RNase H2 in tissue-specific carcinogenesis.
Keywords: Colon Cancer; DNA Repair; Mouse Model; Ribonucleotide Excision Repair.
Copyright © 2019 AGA Institute. Published by Elsevier Inc. All rights reserved.
Figures










References
-
- Arnold M., Sierra M.S., Laversanne M. Global patterns and trends in colorectal cancer incidence and mortality. Gut. 2017;66:683–691. - PubMed
-
- Ferlay J., Soerjomataram I., Dikshit R. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136:E359–E386. - PubMed
-
- Melo FdSe, Kurtova A.V., Harnoss J.M. A distinct role for Lgr5+ stem cells in primary and metastatic colon cancer. Nature. 2017;543:676–680. - PubMed
-
- Tomasetti C., Durrett R., Kimmel M. Role of stem-cell divisions in cancer risk. Nature. 2017;548:E13. - PubMed
Supplementary References
-
- Bartsch K., Knittler K., Borowski C. Absence of RNase H2 triggers generation of immunogenic micronuclei removed by autophagy. Hum Mol Genet. 2017;26:3960–3972. - PubMed
-
- Sato T., Vries R.G., Snippert H.J. Single Lgr5 stem cells build crypt–villus structures in vitro without a mesenchymal niche. Nature. 2009;459:262–265. - PubMed
-
- Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal. 2011;17
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

