DNA G-quadruplex structures mold the DNA methylome
- PMID: 30275516
- PMCID: PMC6173298
- DOI: 10.1038/s41594-018-0131-8
DNA G-quadruplex structures mold the DNA methylome
Abstract
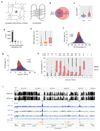
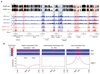
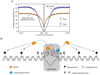
Control of DNA methylation level is critical for gene regulation, and the factors that govern hypomethylation at CpG islands (CGIs) are still being uncovered. Here, we provide evidence that G-quadruplex (G4) DNA secondary structures are genomic features that influence methylation at CGIs. We show that the presence of G4 structure is tightly associated with CGI hypomethylation in the human genome. Surprisingly, we find that these G4 sites are enriched for DNA methyltransferase 1 (DNMT1) occupancy, which is consistent with our biophysical observations that DNMT1 exhibits higher binding affinity for G4s as compared to duplex, hemi-methylated, or single-stranded DNA. The biochemical assays also show that the G4 structure itself, rather than sequence, inhibits DNMT1 enzymatic activity. Based on these data, we propose that G4 formation sequesters DNMT1 thereby protecting certain CGIs from methylation and inhibiting local methylation.
Conflict of interest statement
SB is an advisor and shareholder of Cambridge Epigenetix limited.
Figures

References
-
- Smith ZD, Meissner A. DNA methylation: roles in mammalian development. Nat Rev Genet. 2013;14:204–20. - PubMed
-
- Feinberg AP, Tycko B. The history of cancer epigenetics. Nat Rev Cancer. 2004;4 - PubMed
-
- Li E, Bestor TH, Jaenisch R. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell. 1992;69:915–926. - PubMed
-
- Okano M, Bell DW, Haber DA, Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99:247–257. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

