The role of miR-106p-5p in cervical cancer: from expression to molecular mechanism
- PMID: 30275981
- PMCID: PMC6148547
- DOI: 10.1038/s41420-018-0096-8
The role of miR-106p-5p in cervical cancer: from expression to molecular mechanism
Erratum in
-
Erratum: Publisher Correction: articles initially published in wrong volume.Cell Death Discov. 2019 Jul 10;5:116. doi: 10.1038/s41420-019-0186-2. eCollection 2019. Cell Death Discov. 2019. PMID: 31312525 Free PMC article.
Abstract
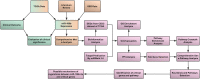
This study aims to investigate the role of miR-106b-5p in cervical cancer by performing a comprehensive analysis on its expression and identifying its putative molecular targets and pathways based on The Cancer Genome Atlas (TCGA) dataset, Gene Expression Omnibus (GEO) dataset, and literature review. Significant upregulation of miR-106b-5p in cervical cancer is confirmed by meta-analysis with the data from TCGA, GEO, and literature. Moreover, the expression of miR-106b-5p is significantly correlated with the number of metastatic lymph nodes. Our bioinformatics analyses show that miR-106b could promote cervical cancer progression by modulating the expression of GSK3B, VEGFA, and PTK2 genes. Importantly, these three genes play a crucial role in PI3K-Akt signaling, focal adhesion, and cancer. Both the expression of miR-106b-5p and key genes are upregulated in cervical cancer. Several explanations could be implemented for this upregulation. However, the specific mechanism needs to be investigated further.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures







References
LinkOut - more resources
Full Text Sources
Miscellaneous

