Misoprostol regulates Bnip3 repression and alternative splicing to control cellular calcium homeostasis during hypoxic stress
- PMID: 30275982
- PMCID: PMC6155004
- DOI: 10.1038/s41420-018-0104-z
Misoprostol regulates Bnip3 repression and alternative splicing to control cellular calcium homeostasis during hypoxic stress
Erratum in
-
Erratum: Publisher Correction: articles initially published in wrong volume.Cell Death Discov. 2019 Jul 10;5:116. doi: 10.1038/s41420-019-0186-2. eCollection 2019. Cell Death Discov. 2019. PMID: 31312525 Free PMC article.
Abstract
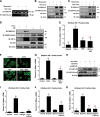
The cellular response to hypoxia involves the activation of a conserved pathway for gene expression regulated by the transcription factor complex called hypoxia-inducible factor (HIF). This pathway has been implicated in both the adaptive response to hypoxia and in several hypoxic-ischemic-related pathologies. Perinatal hypoxic injury, often associated with prematurity, leads to multi-organ dysfunction resulting in significant morbidity and mortality. Using a rodent model of neonatal hypoxia and several representative cell lines, we observed HIF1α activation and down-stream induction of the cell death gene Bnip3 in brain, large intestine, and heart which was mitigated by administration of the prostaglandin E1 analog misoprostol. Mechanistically, we determined that misoprostol inhibits full-length Bnip3 (Bnip3-FL) expression through PKA-mediated NF-κB (P65) nuclear retention, and the induction of pro-survival splice variants. We observed that the dominant small pro-survival variant of Bnip3 in mouse cells lacks the third exon (Bnip3ΔExon3), whereas human cells produce a pro-survival BNIP3 variant lacking exon 2 (BNIP3ΔExon2). In addition, these small Bnip3 splice variants prevent mitochondrial dysfunction, permeability transition, and necrosis triggered by Bnip3-FL by blocking calcium transfer from the sarco/endoplasmic reticulum to the mitochondria. Furthermore, misoprostol and Bnip3ΔExon3 promote nuclear calcium accumulation, resulting in HDAC5 nuclear export, NFAT activation, and adaptive changes in cell morphology and gene expression. Collectively, our data suggests that misoprostol can mitigate the potential damaging effects of hypoxia on multiple cell types by activating adaptive cell survival pathways through Bnip3 repression and alternative splicing.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures







References
LinkOut - more resources
Full Text Sources
Research Materials

