The small protein MgtS and small RNA MgrR modulate the PitA phosphate symporter to boost intracellular magnesium levels
- PMID: 30276893
- PMCID: PMC6351178
- DOI: 10.1111/mmi.14143
The small protein MgtS and small RNA MgrR modulate the PitA phosphate symporter to boost intracellular magnesium levels
Abstract
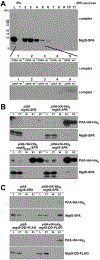
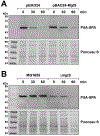
In response to low levels of magnesium (Mg2+ ), the PhoQP two component system induces the transcription of two convergent genes, one encoding a 31-amino acid protein denoted MgtS and the second encoding a small, regulatory RNA (sRNA) denoted MgrR. Previous studies showed that the MgtS protein interacts with and stabilizes the MgtA Mg2+ importer to increase intracellular Mg2+ levels, while the MgrR sRNA base pairs with the eptB mRNA thus affecting lipopolysaccharide modification. Surprisingly, we found overexpression of the MgtS protein also leads to induction of the PhoRB regulon. Studies to understand this activation showed that MgtS forms a complex with a second protein, PitA, a cation-phosphate symporter. Given that the additive effect of ∆mgtA and ∆mgtS mutations on intracellular Mg2+ concentrations seen previously is lost in the ∆pitA mutant, we suggest that MgtS binds to and prevents Mg2+ leakage through PitA under Mg2+ -limiting conditions. Consistent with a detrimental role of PitA in low Mg2+ , we also observe MgrR sRNA repression of PitA synthesis. Thus, PhoQP induces the expression of two convergent small genes in response to Mg2+ limitation whose products act to modulate PitA at different levels to increase intracellular Mg2+ .
© 2018 John Wiley & Sons Ltd.
Figures


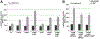
References
-
- Bader MW, Sanowar S, Daley ME, Schneider AR, Cho U, Xu W, Klevit RE, Le Moual H, and Miller SI (2005) Recognition of antimicrobial peptides by a bacterial sensor kinase. Cell 122: 461–472. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

