Transcriptomes of cochlear inner and outer hair cells from adult mice
- PMID: 30277483
- PMCID: PMC6167952
- DOI: 10.1038/sdata.2018.199
Transcriptomes of cochlear inner and outer hair cells from adult mice
Abstract
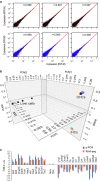
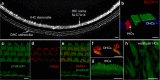
Inner hair cells (IHCs) and outer hair cells (OHCs) are the two anatomically and functionally distinct types of mechanosensitive receptor cells in the mammalian cochlea. The molecular mechanisms defining their morphological and functional specializations are largely unclear. As a first step to uncover the underlying mechanisms, we examined the transcriptomes of IHCs and OHCs isolated from adult CBA/J mouse cochleae. One thousand IHCs and OHCs were separately collected using the suction pipette technique. RNA sequencing of IHCs and OHCs was performed and their transcriptomes were analyzed. The results were validated by comparing some IHC and OHC preferentially expressed genes between present study and published microarray-based data as well as by real-time qPCR. Antibody-based immunocytochemistry was used to validate preferential expression of SLC7A14 and DNM3 in IHCs and OHCs. These data are expected to serve as a highly valuable resource for unraveling the molecular mechanisms underlying different biological properties of IHCs and OHCs as well as to provide a road map for future characterization of genes expressed in IHCs and OHCs.
Conflict of interest statement
The authors declare no competing interests.
Figures


Dataset use reported in
References
Data Citations
References
-
- Hudspeth A. J. Integrating the active process of hair cells with cochlear function. Nat. Rev. Neurosci. 15, 600–614 (2014). - PubMed
-
- Wu Z., Wang X. & Zhang X. Using non-uniform read distribution models to improve isoform expression inference in RNA-Seq. Bioinformatics 27(4), 502–508 (2011). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

