Genome-wide interaction studies identify sex-specific risk alleles for nonsyndromic orofacial clefts
- PMID: 30277614
- PMCID: PMC6185762
- DOI: 10.1002/gepi.22158
Genome-wide interaction studies identify sex-specific risk alleles for nonsyndromic orofacial clefts
Abstract
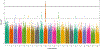
Nonsyndromic cleft lip with or without cleft palate (NSCL/P) is the most common craniofacial birth defect in humans and is notable for its apparent sexual dimorphism where approximately twice as many males are affected as females. The sources of this disparity are largely unknown, but interactions between genetic and sex effects are likely contributors. We examined gene-by-sex (G × S) interactions in a worldwide sample of 2,142 NSCL/P cases and 1,700 controls recruited from 13 countries. First, we performed genome-wide joint tests of the genetic (G) and G × S effects genome-wide using logistic regression assuming an additive genetic model and adjusting for 18 principal components of ancestry. We further interrogated loci with suggestive results from the joint test ( p < 1.00 × 10 -5 ) by examining the G × S effects from the same model. Out of the 133 loci with suggestive results ( p < 1.00 × 10 -5 ) for the joint test, we observed one genome-wide significant G × S effect in the 10q21 locus (rs72804706; p = 6.69 × 10 -9 ; OR = 2.62 CI [1.89, 3.62]) and 16 suggestive G × S effects. At the intergenic 10q21 locus, the risk of NSCL/P is estimated to increase with additional copies of the minor allele for females, but the opposite effect for males. Our observation that the impact of genetic variants on NSCL/P risk differs for males and females may further our understanding of the genetic architecture of NSCL/P and the sex differences underlying clefts and other birth defects.
Keywords: cleft lip; cleft palate; genetic risk; oral facial cleft.
© 2018 Wiley Periodicals, Inc.
Figures

References
-
- Beaty TH, Murray JC, Marazita ML, Munger RG, Ruczinski I, Hetmanski JB, Liang KY, Wu T, Murray T, Fallin MD, Redett RA, Raymond G, Schwender H, Jin SC, Cooper ME, Dunnwald M, Mansilla MA, Leslie E, Bullard S, Lidral AC, Moreno LM, Menezes R, Vieira AR, Petrin A, Wilcox AJ, Lie RT, Jabs EW, Wu-Chou YH, Chen PK, Wang H, Ye X, Huang S, Yeow V, Chong SS, Jee SH, Shi B, Christensen K, Melbye M, Doheny KF, Pugh EW, Ling H, Castilla EE, Czeizel AE, Ma L, Field LL, Brody L, Pangilinan F, Mills JL, Molloy AM, Kirke PN, Scott JM, Arcos-Burgos M, & Scott AF (2010). A genome-wide association study of cleft lip with and without cleft palate identifies risk variants near MAFB and ABCA4. Nat Genet, 42(6), 525–529. 10.1038/ng.580 - DOI - PMC - PubMed
-
- Beaty TH, Ruczinski I, Murray JC, Marazita ML, Munger RG, Hetmanski JB, Murray T, Redett RJ, Fallin MD, Liang KY, Wu T, Patel PJ, Jin SC, Zhang TX, Schwender H, Wu-Chou YH, Chen PK, Chong SS, Cheah F, Yeow V, Ye X, Wang H, Huang S, Jabs EW, Shi B, Wilcox AJ, Lie RT, Jee SH, Christensen K, Doheny KF, Pugh EW, Ling H, & Scott AF (2011). Evidence for gene-environment interaction in a genome wide study of nonsyndromic cleft palate. Genetic Epidemiology, 35(6), 469–478. 10.1002/gepi.20595 - DOI - PMC - PubMed
-
- Beaty TH, Taub MA, Scott AF, Murray JC, Marazita ML, Schwender H, Parker MM, Hetmanski JB, Balakrishnan P, Mansilla MA, Mangold E, Ludwig KU, Noethen MM, Rubini M, Elcioglu N, & Ruczinski I (2013). Confirming genes influencing risk to cleft lip with/without cleft palate in a case-parent trio study. Hum Genet, 132(7), 771–781. 10.1007/s00439-013-1283-6 - DOI - PMC - PubMed
-
- Birnbaum S, Ludwig KU, Reutter H, Herms S, Steffens M, Rubini M, Baluardo C, Ferrian M, Almeida de Assis N, Alblas MA, Barth S, Freudenberg J, Lauster C, Schmidt G, Scheer M, Braumann B, Berge SJ, Reich RH, Schiefke F, Hemprich A, Potzsch S, Steegers-Theunissen RP, Potzsch B, Moebus S, Horsthemke B, Kramer FJ, Wienker TF, Mossey PA, Propping P, Cichon S, Hoffmann P, Knapp M, Nothen MM, & Mangold E (2009). Key susceptibility locus for nonsyndromic cleft lip with or without cleft palate on chromosome 8q24. Nat Genet, 41(4), 473–477. 10.1038/ng.333 - DOI - PubMed
Publication types
MeSH terms
Supplementary concepts
Grants and funding
- R01-DE016148/NH/NIH HHS/United States
- R01 DE014667/DE/NIDCR NIH HHS/United States
- R01-DE014667/NH/NIH HHS/United States
- R01-DE011948/NH/NIH HHS/United States
- K99-DE024571/NH/NIH HHS/United States
- U54 MD007587/MD/NIMHD NIH HHS/United States
- DE011931/NH/NIH HHS/United States
- S21 MD001830/MD/NIMHD NIH HHS/United States
- R01 DE011948/DE/NIDCR NIH HHS/United States
- DD000295/NH/NIH HHS/United States
- U01 DE024425/DE/NIDCR NIH HHS/United States
- DE008559/NH/NIH HHS/United States
- DE009886/NH/NIH HHS/United States
- R00 DE025060/DE/NIDCR NIH HHS/United States
- R01 DE008559/DE/NIDCR NIH HHS/United States
- K99 DE024571/DE/NIDCR NIH HHS/United States
- R01 DD000295/DD/NCBDD CDC HHS/United States
- DE012472/NH/NIH HHS/United States
- U54-MD007587/NH/NIH HHS/United States
- U01-DE024425/NH/NIH HHS/United States
- R37 DE008559/DE/NIDCR NIH HHS/United States
- PICT-2016-3869/Agencia Nacional de Promoción Científica y Tecnológica/International
- R01 DE011931/DE/NIDCR NIH HHS/United States
- R00 DE024571/DE/NIDCR NIH HHS/United States
- DE014667/NH/NIH HHS/United States
- R01 DE016148/DE/NIDCR NIH HHS/United States
- R21 DE016930/DE/NIDCR NIH HHS/United States
- MD001830/NH/NIH HHS/United States
- R00-DE025060/NH/NIH HHS/United States
- R01 DE009886/DE/NIDCR NIH HHS/United States
- X01-HG007485/NH/NIH HHS/United States
- DE016930/NH/NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical

