Substrate Profile of the Phosphotriesterase Homology Protein from Escherichia coli
- PMID: 30277746
- PMCID: PMC6643279
- DOI: 10.1021/acs.biochem.8b00935
Substrate Profile of the Phosphotriesterase Homology Protein from Escherichia coli
Abstract
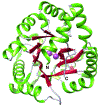
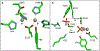
The phosphotriesterase homology protein (PHP) from Escherichia coli is a member of a family of proteins that is related to phosphotriestrase (PTE), a bacterial enzyme from cog1735 with unusual substrate specificity toward the hydrolysis of synthetic organic phosphates and phosphonates. PHP was cloned, purified to homogeneity, and functionally characterized. The three-dimensional structure of PHP was determined at a resolution of 1.84 Å with zinc and phosphate in the active site. The protein folds as a distorted (β/α)8-barrel and possesses a binuclear metal center in the active site. The catalytic function and substrate profile of PHP were investigated using a structure-guided approach that combined bioinformatics, computational docking, organic synthesis, and steady-state enzyme kinetics. PHP was found to catalyze the hydrolysis of phosphorylated glyceryl acetates. The best substrate was 1,2-diacetyl glycerol-3-phosphate with a kcat/ Km of 4.9 × 103 M-1 s-1. The presence of a phosphate group in the substrate was essential for enzymatic hydrolysis by the enzyme. It was surprising, however, to find that PHP was unable to hydrolyze any of the lactones tested as potential substrates, unlike most of the other enzymes from cog1735.
Conflict of interest statement
The authors declare no competing financial interest.
Figures

References
-
- Pieper U, Chiang R, Seffernick JJ, Brown SD, Glasner ME, Kelly L, Eswar N, Sauder JM, Bonanno JB, Swaminathan S, Burley SK, Zheng X, Chance MR, Almo SC, Gerlt JA, Raushel FM, Jacobson MP, Babbitt PC, and Sali A (2009) Target selection and annotation for the structural genomics of the amidohydrolyase and enolase superfamily. J. Struct. Funct. Genomics 10, 107–125. - PMC - PubMed
-
- Wu CH, Aweiler R, Bairoch A, Natale DA, Barker WC, Boeckmann B, Ferro S, Gasteiger E, Huang H, Lopez R, Magrane M, Martin MJ, Mazumander R, O’Donova C, Redaschi N, and Suzek B (2006) The universal protein resource (Uniprot): An expanding universe of protein information. Nucleic Acids Res. 34, D187–D191. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

