PIE-1 Translation in the Germline Lineage Contributes to PIE-1 Asymmetry in the Early Caenorhabditis elegans Embryo
- PMID: 30279189
- PMCID: PMC6288838
- DOI: 10.1534/g3.118.200744
PIE-1 Translation in the Germline Lineage Contributes to PIE-1 Asymmetry in the Early Caenorhabditis elegans Embryo
Abstract
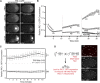
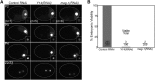
In the C. elegans embryo, the germline lineage is established through successive asymmetric cell divisions that each generate a somatic and a germline daughter cell. PIE-1 is an essential maternal factor that is enriched in embryonic germline cells and is required for germline specification. We estimated the absolute concentration of PIE-1::GFP in germline cells and find that PIE-1::GFP concentration increases by roughly 4.5 fold, from 92 nM to 424 nM, between the 1 and 4-cell stages. Previous studies have shown that the preferential inheritance of PIE-1 by germline daughter cells and the degradation of PIE-1 in somatic cells are important for PIE-1 enrichment in germline cells. In this study, we provide evidence that the preferential translation of maternal PIE-1::GFP transcripts in the germline also contributes to PIE-1::GFP enrichment. Through an RNAi screen, we identified Y14 and MAG-1 (Drosophila tsunagi and mago nashi) as regulators of embryonic PIE-1::GFP levels. We show that Y14 and MAG-1 do not regulate PIE-1 degradation, segregation or synthesis in the early embryo, but do regulate the concentration of maternally-deposited PIE-1::GFP. Taken together, or findings point to an important role for translational control in the regulation of PIE-1 levels in the germline lineage.
Keywords: C. elegans; PIE-1; asymmetric cell division; exon junction complex; germline.
Copyright © 2018 Gauvin et al.
Figures




References
Publication types
MeSH terms
Substances
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
