Chromatin conformation and transcriptional activity are permissive regulators of DNA replication initiation in Drosophila
- PMID: 30279224
- PMCID: PMC6211642
- DOI: 10.1101/gr.239913.118
Chromatin conformation and transcriptional activity are permissive regulators of DNA replication initiation in Drosophila
Abstract
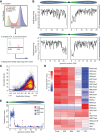
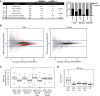
Chromatin structure has emerged as a key contributor to spatial and temporal control over the initiation of DNA replication. However, despite genome-wide correlations between early replication of gene-rich, accessible euchromatin and late replication of gene-poor, inaccessible heterochromatin, a causal relationship between chromatin structure and replication initiation remains elusive. Here, we combined histone gene engineering and whole-genome sequencing in Drosophila to determine how perturbing chromatin structure affects replication initiation. We found that most pericentric heterochromatin remains late replicating in H3K9R mutants, even though H3K9R pericentric heterochromatin is depleted of HP1a, more accessible, and transcriptionally active. These data indicate that HP1a loss, increased chromatin accessibility, and elevated transcription do not result in early replication of heterochromatin. Nevertheless, a small amount of pericentric heterochromatin with increased accessibility replicates earlier in H3K9R mutants. Transcription is de-repressed in these regions of advanced replication but not in those regions of the H3K9R mutant genome that replicate later, suggesting that transcriptional repression may contribute to late replication. We also explored relationships among chromatin, transcription, and replication in euchromatin by analyzing H4K16R mutants. In Drosophila, the X Chromosome gene expression is up-regulated twofold and replicates earlier in XY males than it does in XX females. We found that H4K16R mutation prevents normal male development and abrogates hyperexpression and earlier replication of the male X, consistent with previously established genome-wide correlations between transcription and early replication. In contrast, H4K16R females are viable and fertile, indicating that H4K16 modification is dispensable for genome replication and gene expression.
© 2018 Armstrong et al.; Published by Cold Spring Harbor Laboratory Press.
Figures




References
-
- Aggarwal BD, Calvi BR. 2004. Chromatin regulates origin activity in Drosophila follicle cells. Nature 430: 372–376. - PubMed
-
- Bell SP, Stillman B. 1992. ATP-dependent recognition of eukaryotic origins of DNA replication by a multiprotein complex. Nature 357: 128. - PubMed
-
- Bell O, Schwaiger M, Oakeley EJ, Lienert F, Beisel C, Stadler MB, Schubeler D. 2010. Accessibility of the Drosophila genome discriminates PcG repression, H4K16 acetylation and replication timing. Nat Struct Mol Biol 17: 894–900. - PubMed
-
- Buscaino A, Kocher T, Kind JH, Holz H, Taipale M, Wagner K, Wilm M, Akhtar A. 2003. MOF-regulated acetylation of MSL-3 in the Drosophila dosage compensation complex. Mol Cell 11: 1265–1277. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
