Cyclosporine Biosynthesis in Tolypocladium inflatum Benefits Fungal Adaptation to the Environment
- PMID: 30279281
- PMCID: PMC6168864
- DOI: 10.1128/mBio.01211-18
Cyclosporine Biosynthesis in Tolypocladium inflatum Benefits Fungal Adaptation to the Environment
Abstract
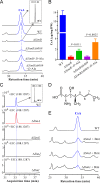
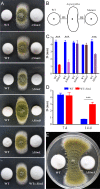
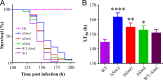
The cycloundecapeptide cyclosporin A (CsA) was first isolated from the insect-pathogenic fungus Tolypocladium inflatum for its antifungal activity and later developed as an immunosuppressant drug. However, the full biosynthetic mechanism of CsA remains unknown and has puzzled researchers for decades. In this study, the biosynthetic gene cluster is suggested to include 12 genes encoding enzymes, including the nonribosomal peptide synthetase (NRPS) (SimA) responsible for assembling the 11 amino acid substrates of cyclosporine and a polyketide synthase (PKS) (SimG) to mediate the production of the unusual amino acid (4R)-4-[(E)-2-butenyl]-4-methyl-l-threonine (Bmt). Individual deletion of 10 genes, isolation of intermediates, and substrate feeding experiments show that Bmt is biosynthesized by three enzymes, including SimG, SimI, and SimJ. The substrate d-alanine is catalyzed from l-alanine by alanine racemase SimB. Gene cluster transcription is regulated by a putative basic leucine zipper (bZIP)-type protein encoded by the cluster gene SimL We also found that the cluster cyclophilin (SimC) and transporter (SimD) genes contribute to the tolerance of CsA in the CsA-producing fungus. We also found that cyclosporine production could enable the fungus to outcompete other fungi during cocultivation tests. Deletion of the CsA biosynthetic genes also impaired fungal virulence against insect hosts. Taking all the data together, in addition to proposing a biosynthetic pathway of cyclosporines, the results of this study suggest that CsA produced by this fungus might play important ecological roles in fungal environment interactions.IMPORTANCE The cyclopeptide cyclosporin A was first isolated from the filamentous fungus Tolypocladium inflatum showing antifungal activity and was later developed as an immunosuppressant drug. We report the biosynthetic mechanism of cyclosporines that are mediated by a cluster of genes encoding NRPS and PKS controlled by a bZIP-type transcriptional regulator. The two unusual amino acids Bmt and d-Ala are produced by the PKS pathway and alanine racemase, respectively. The cyclophilin and transporter genes jointly contribute to fungal self-protection against cyclosporines. Cyclosporine confers on T. inflatum the abilities to outcompete other fungi in competitive interactions and to facilitate fungal infection of insect hosts, which therefore benefits fungal adaptations to different environments.
Keywords: Tolypocladium inflatum; antifungal activity; biosynthetic pathway; cyclosporine; virulence.
Copyright © 2018 Yang et al.
Figures



References
-
- Bushley KE, Raja R, Jaiswal P, Cumbie JS, Nonogaki M, Boyd AE, Owensby CA, Knaus BJ, Elser J, Miller D, Di Y, McPhail KL, Spatafora JW. 2013. The genome of Tolypocladium inflatum: evolution, organization, and expression of the cyclosporin biosynthetic gene cluster. PLoS Genet 9:e1003496. doi:10.1371/journal.pgen.1003496. - DOI - PMC - PubMed
-
- Lawen A, Zocher R. 1990. Cyclosporin synthetase. The most complex peptide synthesizing multienzyme polypeptide so far described. J Biol Chem 265:11355–11360. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

