Conformational changes on substrate binding revealed by structures of Methylobacterium extorquens malate dehydrogenase
- PMID: 30279311
- PMCID: PMC6168771
- DOI: 10.1107/S2053230X18011809
Conformational changes on substrate binding revealed by structures of Methylobacterium extorquens malate dehydrogenase
Abstract
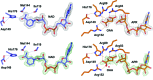
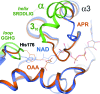
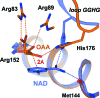
Three high-resolution X-ray crystal structures of malate dehydrogenase (MDH; EC 1.1.1.37) from the methylotroph Methylobacterium extorquens AM1 are presented. By comparing the structures of apo MDH, a binary complex of MDH and NAD+, and a ternary complex of MDH and oxaloacetate with ADP-ribose occupying the pyridine nucleotide-binding site, conformational changes associated with the formation of the catalytic complex were characterized. While the substrate-binding site is accessible in the enzyme resting state or NAD+-bound forms, the substrate-bound form exhibits a closed conformation. This conformational change involves the transition of an α-helix to a 310-helix, which causes the adjacent loop to close the active site following coenzyme and substrate binding. In the ternary complex, His284 forms a hydrogen bond to the C2 carbonyl of oxaloacetate, placing it in a position to donate a proton in the formation of (2S)-malate.
Keywords: Methylobacterium extorquens; biofuels; malate dehydrogenase; methylotrophs.
Figures
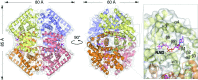
Similar articles
-
Structure of glyoxysomal malate dehydrogenase (MDH3) from Saccharomyces cerevisiae.Acta Crystallogr F Struct Biol Commun. 2018 Oct 1;74(Pt 10):617-624. doi: 10.1107/S2053230X18011895. Epub 2018 Sep 19. Acta Crystallogr F Struct Biol Commun. 2018. PMID: 30279312 Free PMC article.
-
Structure of Methylobacterium extorquens malyl-CoA lyase: CoA-substrate binding correlates with domain shift.Acta Crystallogr F Struct Biol Commun. 2017 Feb 1;73(Pt 2):79-85. doi: 10.1107/S2053230X17001029. Epub 2017 Jan 27. Acta Crystallogr F Struct Biol Commun. 2017. PMID: 28177317 Free PMC article.
-
An unusual diphosphatase from the PhnP family cleaves reactive FAD photoproducts.Biochem J. 2018 Jan 11;475(1):261-272. doi: 10.1042/BCJ20170817. Biochem J. 2018. PMID: 29229761
-
Structural Comparison of hMDH2 Complexed with Natural Substrates and Cofactors: The Importance of Phosphate Binding for Active Conformation and Catalysis.Biomolecules. 2022 Aug 25;12(9):1175. doi: 10.3390/biom12091175. Biomolecules. 2022. PMID: 36139014 Free PMC article.
-
Catalytic mechanism and kinetics of malate dehydrogenase.Essays Biochem. 2024 Oct 3;68(2):73-82. doi: 10.1042/EBC20230086. Essays Biochem. 2024. PMID: 38721782 Free PMC article. Review.
Cited by
-
Protein Conformational Space at the Edge of Allostery: Turning a Nonallosteric Malate Dehydrogenase into an "Allosterized" Enzyme Using Evolution-Guided Punctual Mutations.Mol Biol Evol. 2022 Sep 1;39(9):msac186. doi: 10.1093/molbev/msac186. Mol Biol Evol. 2022. PMID: 36056899 Free PMC article.
References
-
- Anthony, C. (1982). The Biochemistry of Methylotrophs. London: Academic Press.
-
- Bhat, T. N. (1988). J. Appl. Cryst. 21, 279–281.
-
- Chapman, A. D. M., Cortés, A., Dafforn, T. R., Clarke, A. R. & Brady, R. L. (1999). J. Mol. Biol. 285, 703–712. - PubMed
-
- Colowick, S. P. & Kaplan, N. O. (1957). Methods Enzymol. 4, 840–855.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

