Cationic domains in particle-forming and assembly-deficient HBV core antigens capture mammalian RNA that stimulates Th1-biased antibody responses by DNA vaccination
- PMID: 30279478
- PMCID: PMC6168482
- DOI: 10.1038/s41598-018-32971-5
Cationic domains in particle-forming and assembly-deficient HBV core antigens capture mammalian RNA that stimulates Th1-biased antibody responses by DNA vaccination
Abstract
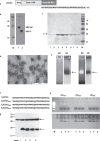
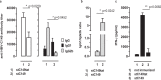
The HBV core protein self-assembles into particles and encapsidates immune-stimulatory bacterial RNA through a cationic COOH-terminal (C150-183) domain. To investigate if different cationic domains have an impact on the endogenous RNA-binding of HBV-C antigens in mammalian cells, we developed a strep-tag (st) based expression/purification system for HBV-C/RNA antigens in vector-transfected HEK-293 cells. We showed that HBV-stC but not HBV-stC149 particles (lacking the cationic domain) capture low amounts of mammalian RNA. Prevention of specific phosphorylation in cationic domains, either by exchanging the serine residues S155, S162 and S170 with alanines (HBV-stCAAA) or by exchanging the entire cationic domain with a HIV-tat48-57-like sequence (HBV-stC149tat) enhanced the encapsidation of RNA into mutant core particles. Particle-bound mammalian RNA functioned as TLR-7 ligand and induced a Th1-biased humoral immunity in B6 but not in TLR-7-/- mice by exogenous (protein) and endogenous (DNA) vaccines. Compared to core particles, binding of mammalian RNA to freely exposed cationic domains in assembly-deficient antigens was enhanced. However, RNA bound to non-particulate antigens unleash its Th1-stimulating adjuvant activity by DNA- but not protein-based vaccination. Mammalian RNAs targeted by an endogenously expressed antigen thus function as a natural adjuvant in the host that facilitates priming of Th1-biased immune responses by DNA-based immunization.
Conflict of interest statement
The authors declare no competing interests.
Figures



Similar articles
-
Endogenously Expressed Antigens Bind Mammalian RNA via Cationic Domains that Enhance Priming of Effector CD8 T Cells by DNA Vaccination.Mol Ther. 2019 Mar 6;27(3):661-672. doi: 10.1016/j.ymthe.2019.01.011. Epub 2019 Jan 22. Mol Ther. 2019. PMID: 30713086 Free PMC article.
-
Priming Th1 immunity to viral core particles is facilitated by trace amounts of RNA bound to its arginine-rich domain.J Immunol. 2002 May 15;168(10):4951-9. doi: 10.4049/jimmunol.168.10.4951. J Immunol. 2002. PMID: 11994446
-
Expression and immunoactivity of chimeric particulate antigens of receptor binding site-core antigen of hepatitis B virus.World J Gastroenterol. 2005 Jan 28;11(4):492-7. doi: 10.3748/wjg.v11.i4.492. World J Gastroenterol. 2005. PMID: 15641132 Free PMC article.
-
DNA-mediated immunization to the hepatitis B surface antigen. Activation and entrainment of the immune response.Ann N Y Acad Sci. 1995 Nov 27;772:64-76. doi: 10.1111/j.1749-6632.1995.tb44732.x. Ann N Y Acad Sci. 1995. PMID: 8546414 Review.
-
Hybrid hepatitis B virus core antigen as a vaccine carrier moiety: I. presentation of foreign epitopes.J Biotechnol. 1996 Jan 26;44(1-3):91-6. doi: 10.1016/0168-1656(95)00118-2. J Biotechnol. 1996. PMID: 8717391 Review.
Cited by
-
PreS1 Containing HBc VLPs for the Development of a Combined Therapeutic/Prophylactic Hepatitis B Vaccine.Microorganisms. 2023 Apr 8;11(4):972. doi: 10.3390/microorganisms11040972. Microorganisms. 2023. PMID: 37110395 Free PMC article.
-
Endogenously Expressed Antigens Bind Mammalian RNA via Cationic Domains that Enhance Priming of Effector CD8 T Cells by DNA Vaccination.Mol Ther. 2019 Mar 6;27(3):661-672. doi: 10.1016/j.ymthe.2019.01.011. Epub 2019 Jan 22. Mol Ther. 2019. PMID: 30713086 Free PMC article.
-
Introducing adjuvant-loaded particulate hepatitis B core antigen as an alternative therapeutic hepatitis B vaccine component.JHEP Rep. 2023 Dec 30;6(4):100997. doi: 10.1016/j.jhepr.2023.100997. eCollection 2024 Apr. JHEP Rep. 2023. PMID: 38425450 Free PMC article.
-
Heterologous DNA-prime/protein-boost immunization with a monomeric SARS-CoV-2 spike antigen redundantizes the trimeric receptor-binding domain structure to induce neutralizing antibodies in old mice.Front Immunol. 2023 Sep 11;14:1231274. doi: 10.3389/fimmu.2023.1231274. eCollection 2023. Front Immunol. 2023. PMID: 37753087 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

