Megahertz serial crystallography
- PMID: 30279492
- PMCID: PMC6168542
- DOI: 10.1038/s41467-018-06156-7
Megahertz serial crystallography
Abstract
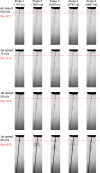
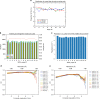
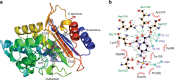
The new European X-ray Free-Electron Laser is the first X-ray free-electron laser capable of delivering X-ray pulses with a megahertz inter-pulse spacing, more than four orders of magnitude higher than previously possible. However, to date, it has been unclear whether it would indeed be possible to measure high-quality diffraction data at megahertz pulse repetition rates. Here, we show that high-quality structures can indeed be obtained using currently available operating conditions at the European XFEL. We present two complete data sets, one from the well-known model system lysozyme and the other from a so far unknown complex of a β-lactamase from K. pneumoniae involved in antibiotic resistance. This result opens up megahertz serial femtosecond crystallography (SFX) as a tool for reliable structure determination, substrate screening and the efficient measurement of the evolution and dynamics of molecular structures using megahertz repetition rate pulses available at this new class of X-ray laser source.
Conflict of interest statement
The authors declare no competing interests.
Figures


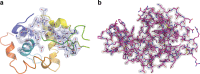
References
Publication types
Grants and funding
- Project oriented funds/Helmholtz-Gemeinschaft (Helmholtz Gemeinschaft)/International
- 609920/ERC_/European Research Council/International
- WT_/Wellcome Trust/United Kingdom
- DFG-EXC1074/Deutsche Forschungsgemeinschaft (German Research Foundation)/International
- R01 GM117342/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources

