How phosphorylation influences E1 subunit pyruvate dehydrogenase: A computational study
- PMID: 30279533
- PMCID: PMC6168537
- DOI: 10.1038/s41598-018-33048-z
How phosphorylation influences E1 subunit pyruvate dehydrogenase: A computational study
Abstract
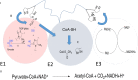
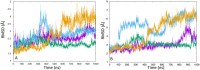
Pyruvate (PYR) dehydrogenase complex (PDC) is an enzymatic system that plays a crucial role in cellular metabolism as it controls the entry of carbon into the Krebs cycle. From a structural point of view, PDC is formed by three different subunits (E1, E2 and E3) capable of catalyzing the three reaction steps necessary for the full conversion of pyruvate to acetyl-CoA. Recent investigations pointed out the crucial role of this enzyme in the replication and survival of specific cancer cell lines, renewing the interest of the scientific community. Here, we report the results of our molecular dynamics studies on the mechanism by which posttranslational modifications, in particular the phosphorylation of three serine residues (Ser-264-α, Ser-271-α, and Ser-203-α), influence the enzymatic function of the protein. Our results support the hypothesis that the phosphorylation of Ser-264-α and Ser-271-α leads to (1) a perturbation of the catalytic site structure and dynamics and, especially in the case of Ser-264-α, to (2) a reduction in the affinity of E1 for the substrate. Additionally, an analysis of the channels connecting the external environment with the catalytic site indicates that the inhibitory effect should not be due to the occlusion of the access/egress pathways to/from the active site.
Conflict of interest statement
The authors declare no competing interests.
Figures




References
-
- He H, Xia H, Xia Q, Ren Y, He H. Design and optimization of N-acylhydrazone pyrimidine derivatives as E. coli PDHc E1 inhibitors: Structure-activity relationship analysis, biological evaluation and molecular docking study. Bioorg. Med. Chem. 2017;25:5652-. doi: 10.1016/j.bmc.2017.08.038. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

