Differing isoforms of the cobalamin binding photoreceptor AerR oppositely regulate photosystem expression
- PMID: 30281022
- PMCID: PMC6199135
- DOI: 10.7554/eLife.39028
Differing isoforms of the cobalamin binding photoreceptor AerR oppositely regulate photosystem expression
Abstract
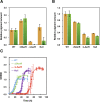
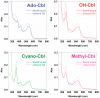
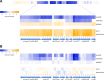
Phototrophic microorganisms adjust photosystem synthesis in response to changes in light intensity and wavelength. A variety of different photoreceptors regulate this process. Purple photosynthetic bacteria synthesize a novel photoreceptor AerR that uses cobalamin (B12) as a blue-light absorbing chromophore to control photosystem synthesis. AerR directly interacts with the redox responding transcription factor CrtJ, affecting CrtJ's interaction with photosystem promoters. In this study, we show that AerR is translated as two isoforms that differ by 41 amino acids at the amino terminus. The ratio of these isoforms was affected by light and cell growth phase with the long variant predominating during photosynthetic exponential growth and the short variant predominating in dark conditions and/or stationary phase. Pigmentation and transcriptomic analyses show that the short AerR variant represses, while long variant activates, photosynthesis genes. The long form of AerR also activates many genes involved in cellular metabolism and motility.
Keywords: B12 photoreceptor; CrtJ/PpsR; Rhodobacter capsulatus; anoxygenic photosynthesis; genetics; genomics.
© 2018, Yamamoto et al.
Conflict of interest statement
HY, MF, VD, CB No competing interests declared
Figures













References
-
- Armstrong GA. Genetic Analysis and Regulation of Carotenoid Biosynthesis. In: Blankenship R. E, Madigan M. T, Bauer C. E, editors. Anoxygenic Photosynthetic Bacteria. Dordrecht: Springer Netherlands; 1995. pp. 1135–1157.
-
- Babic S, Hunter CN, Rakhlin NJ, Simons RW, Phillips-Jones MK. Molecular characterisation of the pifC gene encoding translation initiation factor 3, which is required for normal photosynthetic complex formation in Rhodobacter sphaeroides NCIB 8253. European Journal of Biochemistry. 1997;249:564–575. doi: 10.1111/j.1432-1033.1997.t01-1-00564.x. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

