Genomic and transcriptomic characterisation of undifferentiated pleomorphic sarcoma of bone
- PMID: 30281149
- PMCID: PMC8033574
- DOI: 10.1002/path.5176
Genomic and transcriptomic characterisation of undifferentiated pleomorphic sarcoma of bone
Abstract
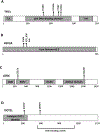
Undifferentiated pleomorphic sarcoma of bone (UPSb) is a rare primary bone sarcoma that lacks a specific line of differentiation. There is very little information about the genetic alterations leading to tumourigenesis or malignant transformation. Distinguishing between UPSb and other malignant bone sarcomas, including dedifferentiated chondrosarcoma and osteosarcoma, can be challenging due to overlapping features. To explore the genomic and transcriptomic landscape of UPSb tumours, whole-exome sequencing (WES) and RNA sequencing (RNA-Seq) were performed on UPSb tumours. All tumours lacked hotspot mutations in IDH1/2 132 or 172 codons, thereby excluding the diagnosis of dedifferentiated chondrosarcoma. Recurrent somatic mutations in TP53 were identified in four of 14 samples (29%). Moreover, recurrent mutations in histone chromatin remodelling genes, including H3F3A, ATRX and DOT1L, were identified in five of 14 samples (36%), highlighting the potential role of deregulated chromatin remodelling pathways in UPSb tumourigenesis. The majority of recurrent mutations in chromatin remodelling genes identified here are reported in COSMIC, including the H3F3A G34 and K36 hotspot residues. Copy number alteration analysis identified gains and losses in genes that have been previously altered in UPSb or UPS of soft tissue. Eight somatic gene fusions were identified by RNA-Seq, two of which, CLTC-VMP1 and FARP1-STK24, were reported previously in multiple cancers. Five gene fusions were genomically characterised. Hierarchical clustering analysis, using RNA-Seq data, distinctly clustered UPSb tumours from osteosarcoma and other sarcomas, thus molecularly distinguishing UPSb from other sarcomas. RNA-Seq expression profiling analysis and quantitative reverse transcription-polymerase chain reaction showed an elevated expression in FGF23, which can be a potential molecular biomarker for UPSb. To our knowledge, this study represents the first comprehensive WES and RNA-Seq analysis of UPSb tumours revealing novel protein-coding recurrent gene mutations, gene fusions and identifying a potential UPSb molecular biomarker, thereby broadening the understanding of the pathogenic mechanisms and highlighting the possibility of developing novel targeted therapeutics. Copyright © 2018 Pathological Society of Great Britain and Ireland. Published by John Wiley & Sons, Ltd.
Keywords: CNV; FGF23; RNA sequencing; Whole exome sequencing; chromatin remodelling genes; gene expression and hierarchical clustering analyses; gene fusions; sarcomas; undifferentiated pleomorphic sarcoma of bone.
Copyright © 2018 Pathological Society of Great Britain and Ireland. Published by John Wiley & Sons, Ltd.
Conflict of interest statement
Figures



References
-
- Fletcher CDM. WHO classification of tumours of soft tissue and bone. 4th edition. International Agency for Research on Cancer, Lyon: 2013.
-
- Niini T, Lahti L, Michelacci F, et al. Array comparative genomic hybridization reveals frequent alterations of G1/S checkpoint genes in undifferentiated pleomorphic sarcoma of bone. Genes Chromosomes Cancer 2011; 50: 291–306. - PubMed
-
- Doyle LA. Sarcoma classification: an update based on the 2013 World Health Organization Classification of Tumors of Soft Tissue and Bone. Cancer 2014; 120: 1763–1774. - PubMed
-
- Chen S, Fritchie K, Wei S, et al. Diagnostic utility of IDH1/2 mutations to distinguish dedifferentiated chondrosarcoma from undifferentiated pleomorphic sarcoma of bone. Hum Pathol 2017; 65: 239–246. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

